8GJF
 
 | | afupcna bound with peptide mimetic | | Descriptor: | LYS-ARG-ARG-GLN-THR-SER-MET-THR-ASP-PHE-TYR-HIS-SER-LYS-ARG, Proliferating cell nuclear antigen | | Authors: | Vandborg, B, Bruning, J.B. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Towards a High-Affinity Peptidomimetic Targeting Proliferating Cell Nuclear Antigen from Aspergillus fumigatus.
J Fungi, 9, 2023
|
|
8GC8
 
 | |
8GC7
 
 | |
8GA2
 
 | | Bromodomain of CBP liganded with inhibitor iCBP5 | | Descriptor: | (6S)-6-{(5M)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}-1-phenylpiperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8GET
 
 | | R. hominis 2 beta-glucuronidase bound to norquetiapine-glucuronide | | Descriptor: | 11-(4-beta-D-glucopyranuronosylpiperazin-1-yl)dibenzo[b,f][1,4]thiazepine, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
2C6I
 
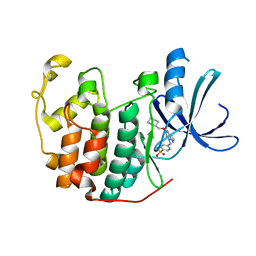 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | Descriptor: | 4-{[5-(CYCLOHEXYLMETHOXY)[1,2,4]TRIAZOLO[1,5-A]PYRIMIDIN-7-YL]AMINO}BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2005-11-10 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Triazolo[1,5-A]Pyrimidines as Novel Cdk2 Inhibitors: Protein Structure-Guided Design and Sar.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2C6M
 
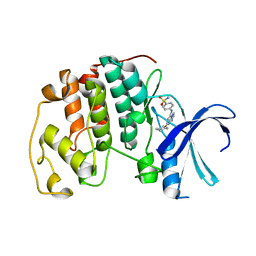 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | Descriptor: | 4-{[5-(CYCLOHEXYLOXY)[1,2,4]TRIAZOLO[1,5-A]PYRIMIDIN-7-YL]AMINO}BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2005-11-10 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Triazolo[1,5-A]Pyrimidines as Novel Cdk2 Inhibitors: Protein Structure-Guided Design and Sar.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2XDE
 
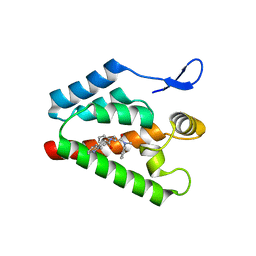 | | Crystal structure of the complex of PF-3450074 with an engineered HIV capsid N terminal domain | | Descriptor: | GAG POLYPROTEIN, N-METHYL-NALPHA-[(2-METHYL-1H-INDOL-3-YL)ACETYL]-N-PHENYL-L-PHENYLALANINAMIDE | | Authors: | Brown, D.G, Irving, S.L, Anderson, M, Bazin, R. | | Deposit date: | 2010-04-30 | | Release date: | 2010-12-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | HIV Capsid is a Tractable Target for Small Molecule Therapeutic Intervention.
Plos Pathog., 6, 2010
|
|
8FMI
 
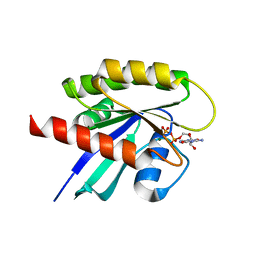 | | Crystal structure of human KRAS at 1.12 A | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION | | Authors: | Brenner, R, Landgraf, A, Gonzalez-Gutierrez, G, Bum-Erdene, K, Meroueh, S.O. | | Deposit date: | 2022-12-23 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Packing Reveals a Potential Autoinhibited KRAS Dimer Interface and a Strategy for Small-Molecule Inhibition of RAS Signaling.
Biochemistry, 62, 2023
|
|
8FMJ
 
 | | Crystal structure of human KRAS in space group R32 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Brenner, R, Landgraf, A, Gonzalez-Gutierrez, G, Bum-Erdene, K, Meroueh, S.O. | | Deposit date: | 2022-12-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal Packing Reveals a Potential Autoinhibited KRAS Dimer Interface and a Strategy for Small-Molecule Inhibition of RAS Signaling.
Biochemistry, 62, 2023
|
|
8FTN
 
 | | E. coli ArnA dehydrogenase domain mutant - N492A | | Descriptor: | Bifunctional UDP-4-amino-4-deoxy-L-arabinose formyltransferase/UDP-glucuronic acid oxidase ArnA, SULFATE ION | | Authors: | Sousa, M.C, Mitchell, M.E. | | Deposit date: | 2023-01-12 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeting the Conformational Change in ArnA Dehydrogenase for Selective Inhibition of Polymyxin Resistance.
Biochemistry, 62, 2023
|
|
8FMK
 
 | | Crystal structure of human KRAS with extended switch I loop | | Descriptor: | CHLORIDE ION, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Brenner, R, Landgraf, A, Gonzalez-Gutierrez, G, Bum-Erdene, K, Meroueh, S.O. | | Deposit date: | 2022-12-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Packing Reveals a Potential Autoinhibited KRAS Dimer Interface and a Strategy for Small-Molecule Inhibition of RAS Signaling.
Biochemistry, 62, 2023
|
|
8GEO
 
 | | E. eligens beta-glucuronidase bound to 3-OH-desloratidine-glucuronide | | Descriptor: | 8-chloro-11-(1-beta-D-glucopyranuronosylpiperidin-4-ylidene)-3-hydroxy-6,11-dihydro-5H-benzo[5,6]cyclohepta[1,2-b]pyridine, Beta-glucuronidase, GLYCEROL | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
2V1M
 
 | | Crystal structure of Schistosoma mansoni glutathione peroxidase | | Descriptor: | ACETATE ION, GLUTATHIONE PEROXIDASE, LITHIUM ION, ... | | Authors: | Dimastrogiovanni, D, Miele, A.E, Angelucci, F, Boumis, G, Bellelli, A, Brunori, M. | | Deposit date: | 2008-09-16 | | Release date: | 2009-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Combining Crystallography and Molecular Dynamics: The Case of Schistosoma Mansoni Phospholipid Glutathione Peroxidase.
Proteins, 78, 2010
|
|
2WHX
 
 | | A second conformation of the NS3 protease-helicase from dengue virus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Luo, D, Lescar, J. | | Deposit date: | 2009-05-07 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Flexibility between the Protease and Helicase Domains of the Dengue Virus Ns3 Protein Conferred by the Linker Region and its Functional Implications.
J.Biol.Chem., 285, 2010
|
|
8FVS
 
 | | Bromodomain of CBP liganded with CCS1477int | | Descriptor: | 1,2-ETHANEDIOL, Histone acetyltransferase, MAGNESIUM ION, ... | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
2XXS
 
 | | Solution structure of the N-terminal domain of the Shigella type III secretion protein MxiG | | Descriptor: | PROTEIN MXIG | | Authors: | McDowell, M.A, Johnson, S, Deane, J.E, McDonnell, J.M, Lea, S.M. | | Deposit date: | 2010-11-11 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Studies on the N-Terminal Domain of the Shigella Type III Secretion Protein Mxig.
J.Biol.Chem., 286, 2011
|
|
2WZQ
 
 | | Insertion Mutant E173GP174 of the NS3 protease-helicase from dengue virus | | Descriptor: | CHLORIDE ION, GLYCEROL, NS3 PROTEASE-HELICASE | | Authors: | Luo, D, Wei, N, Doan, D, Paradkar, P, Chong, Y, Davidson, A, Kotaka, M, Lescar, J, Vasudevan, S. | | Deposit date: | 2009-12-02 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Flexibility between the Protease and Helicase Domains of the Dengue Virus Ns3 Protein Conferred by the Linker Region and its Functional Implications.
J.Biol.Chem., 285, 2010
|
|
2AS5
 
 | | Structure of the DNA binding domains of NFAT and FOXP2 bound specifically to DNA. | | Descriptor: | 5'-D(AP*AP*CP*TP*AP*TP*GP*AP*AP*AP*CP*AP*AP*AP*TP*TP*TP*TP*CP*CP*TP*)-3', 5'-D(TP*TP*AP*GP*GP*AP*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*CP*AP*TP*AP*GP*)-3', Forkhead box protein P2, ... | | Authors: | Wu, Y, Stroud, J.C, Borde, M, Bates, D.L, Guo, L, Han, A, Rao, A, Chen, L. | | Deposit date: | 2005-08-22 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | FOXP3 Controls Regulatory T Cell Function through Cooperation with NFAT.
Cell(Cambridge,Mass.), 126, 2006
|
|
2CU9
 
 | |
5A5O
 
 | | Crystal structure of human ATAD2 bromodomain in complex with 3-methyl- 1,2-dihydroquinolin-2-one | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYL-1,2-DIHYDROQUINOLIN-2-ONE, ATPASE FAMILY AAA DOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2015-06-20 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Fragment-Based Discovery of Low-Micromolar Atad2 Bromodomain Inhibitors.
J.Med.Chem., 58, 2015
|
|
5BZ3
 
 | | CRYSTAL STRUCTURE OF SODIUM PROTON ANTIPORTER NAPA IN OUTWARD-FACING CONFORMATION. | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL (7Z)-TETRADEC-7-ENOATE, Na(+)/H(+) antiporter | | Authors: | Coincon, M, Uzdavinys, P, Emmanuel, N, Cameron, A, Drew, D. | | Deposit date: | 2015-06-11 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures reveal the molecular basis of ion translocation in sodium/proton antiporters.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5C9J
 
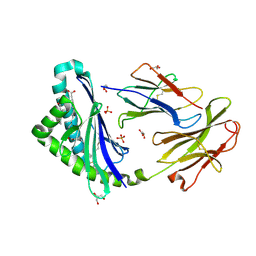 | | Human CD1c with ligands in A' and F' channel | | Descriptor: | Beta-2-microglobulin, GLYCEROL, LAURIC ACID, ... | | Authors: | Tews, I, Machelett, M.M, Mansour, S, Gadola, S.D. | | Deposit date: | 2015-06-27 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cholesteryl esters stabilize human CD1c conformations for recognition by self-reactive T cells.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5BZ2
 
 | |
1O80
 
 | |
