3IDF
 
 | |
3IS6
 
 | | The Crystal Structure of a domain of a putative Permease protein from Porphyromonas gingivalis to 2A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, putative permease protein, ... | | Authors: | Stein, A.J, Sather, A, Duggan, E, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-25 | | Release date: | 2009-09-08 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of a domain of a putative Permease protein from Porphyromonas gingivalis to 2A
To be Published
|
|
4MC9
 
 | | HIV protease in complex with AA74 | | Descriptor: | (3S)-tetrahydrofuran-3-yl {(2S,3R)-4-[(4R)-7-fluoro-1,1-dioxido-4-(propan-2-yl)-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}carbamate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Ganguly, A.K, Alluri, S.S, Wang, C, Antropow, A, White, A, Caroccia, D, Biswas, D, Kang, E, Zhang, L, Carroll, S.S, Burlein, C, Munshi, V, Orth, P, Strickland, C. | | Deposit date: | 2013-08-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural Optimization of Cyclic Sulfonamide based Novel HIV-1 Protease Inhibitors to Pico Molar Affinities guided by X-ray Crystallographic Analysis
Tetrahedron, 2014
|
|
3NW2
 
 | | Novel nanomolar Imidazopyridines as selective Nitric Oxide Synthase (iNOS) inhibitors: SAR and structural insights | | Descriptor: | 2-[2-(4-methoxypyridin-2-yl)ethyl]-3H-imidazo[4,5-b]pyridine, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Graedler, U, Fuchss, T, Ulrich, W.R, Boer, R, Strub, A, Hesslinger, C, Anezo, C, Diederichs, K, Zaliani, A. | | Deposit date: | 2010-07-09 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel nanomolar imidazo[4,5-b]pyridines as selective nitric oxide synthase (iNOS) inhibitors: SAR and structural insights
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1YKV
 
 | | Crystal structure of the Diels-Alder ribozyme complexed with the product of the reaction between N-pentylmaleimide and covalently attached 9-hydroxymethylanthracene | | Descriptor: | (3AS,9AS)-2-PENTYL-4-HYDROXYMETHYL-3A,4,9,9A-TETRAHYDRO-4,9[1',2']-BENZENO-1H-BENZ[F]ISOINDOLE-1,3(2H)-DIONE, Diels-Alder ribozyme, MAGNESIUM ION | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
4MD1
 
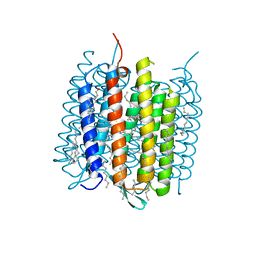 | | Orange species of bacteriorhodopsin from Halobacterium salinarum | | Descriptor: | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Borshchevskiy, V, Erofeev, I, Round, E, Weik, M, Ishchenko, A, Gushchin, I, Mishin, A, Bueldt, G, Gordeliy, V. | | Deposit date: | 2013-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Low-dose X-ray radiation induces structural alterations in proteins.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4H8I
 
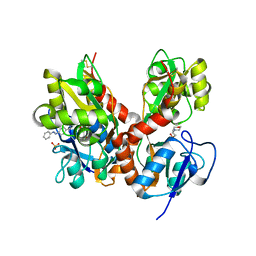 | | Structure of GluK2-LBD in complex with GluAzo | | Descriptor: | (4R)-4-[(2E)-3-{4-[(E)-phenyldiazenyl]phenyl}prop-2-en-1-yl]-L-glutamic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Reiter, A, Skerra, A, Trauner, D, Schiefner, A. | | Deposit date: | 2012-09-22 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A photoswitchable neurotransmitter analogue bound to its receptor.
Biochemistry, 52, 2013
|
|
5EVO
 
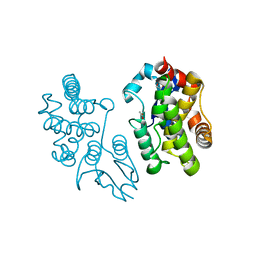 | | Structure of Dehydroascrobate Reductase from Pennisetum Americanum in complex with two non-native ligands, Acetate in the G-site and Glycerol in the H-site | | Descriptor: | ACETATE ION, Dehydroascorbate reductase, GLYCEROL | | Authors: | Kumar, A.O, Das, B.K, Arockiasamy, A. | | Deposit date: | 2015-11-20 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Non-native ligands define the active site of Pennisetum glaucum (L.) R. Br dehydroascorbate reductase.
Biochem.Biophys.Res.Commun., 473, 2016
|
|
6HI5
 
 | | The ATAD2 bromodomain in complex with compound 8 | | Descriptor: | (2~{R})-2-azanyl-~{N}-[5-(5-azanylpyridin-3-yl)-4-ethanoyl-1,3-thiazol-2-yl]propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2018-08-29 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
6HIA
 
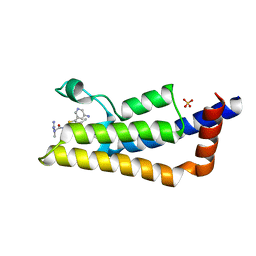 | | The ATAD2 bromodomain in complex with compound 13 | | Descriptor: | (2~{R})-~{N}-[5-(5-azanylpyridin-3-yl)-4-ethanoyl-1,3-thiazol-2-yl]-2-carbamimidamido-propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2018-08-29 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
6HOD
 
 | |
5F4E
 
 | | Crystal structure of the human sperm Izumo1 and egg Juno complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Aydin, H, Sultana, A, Lee, J.E. | | Deposit date: | 2015-12-03 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular architecture of the human sperm IZUMO1 and egg JUNO fertilization complex.
Nature, 534, 2016
|
|
5F4V
 
 | |
4D7A
 
 | | Crystal structure of E. coli tRNA N6-threonylcarbamoyladenosine dehydratase, TcdA, in complex with AMP at 1.801 Angstroem resolution | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lopez-Estepa, M, Arda, A, Savko, M, Round, A, Shepard, W, Bruix, M, Coll, M, Fernandez, F.J, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2014-11-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The Crystal Structure and Small-Angle X-Ray Analysis of Csdl/Tcda Reveal a New tRNA Binding Motif in the Moeb/E1 Superfamily.
Plos One, 10, 2015
|
|
3IPU
 
 | | X-ray structure of benzisoxazole urea synthetic agonist bound to the LXR-alpha | | Descriptor: | 4-{[methyl(3-{[7-propyl-3-(trifluoromethyl)-1,2-benzisoxazol-6-yl]oxy}propyl)carbamoyl]amino}benzoic acid, Nuclear receptor coactivator 1, Oxysterols receptor LXR-alpha, ... | | Authors: | Fradera, X, Vu, D, Nimz, O, Skene, R, Hosfield, D, Wijnands, R, Cooke, A.J, Haunso, A, King, A, Bennet, D.J, McGuire, R, Uitdehaag, J.C.M. | | Deposit date: | 2009-08-18 | | Release date: | 2010-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of the LXRalpha LBD in its homodimeric form and implications for heterodimer signaling.
J.Mol.Biol., 399, 2010
|
|
5F5W
 
 | |
3I7M
 
 | | N-terminal domain of Xaa-Pro dipeptidase from Lactobacillus brevis. | | Descriptor: | Xaa-Pro dipeptidase | | Authors: | Osipiuk, J, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-08 | | Release date: | 2009-07-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | X-ray crystal structure of N-terminal domain of Xaa-Pro dipeptidase from Lactobacillus brevis.
To be Published
|
|
7WPH
 
 | | SARS-CoV2 RBD bound to Fab06 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB06 light chain, Fab06 heavy chain, ... | | Authors: | Lin, J.Q, El Sahili, A, Lescar, J. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Engineering SARS-CoV-2 specific cocktail antibodies into a bispecific format improves neutralizing potency and breadth.
Nat Commun, 13, 2022
|
|
5EW9
 
 | | Crystal Structure of Aurora A Kinase Domain Bound to MK-5108 | | Descriptor: | 4-(3-chloranyl-2-fluoranyl-phenoxy)-1-[[6-(1,3-thiazol-2-ylamino)pyridin-2-yl]methyl]cyclohexane-1-carboxylic acid, Aurora kinase A | | Authors: | Shiau, A.K, Motamedi, A. | | Deposit date: | 2015-11-20 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | A Cell Biologist's Field Guide to Aurora Kinase Inhibitors.
Front Oncol, 5, 2015
|
|
7WPV
 
 | | Fab14 - a SARS-CoV2 RBD neutralising antibody | | Descriptor: | Fab14 heavy chain, Fab14 light chain | | Authors: | Lin, J.Q, El Sahili, A, Lescar, J. | | Deposit date: | 2022-01-24 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Engineering SARS-CoV-2 specific cocktail antibodies into a bispecific format improves neutralizing potency and breadth.
Nat Commun, 13, 2022
|
|
3O5B
 
 | | Crystal structure of dimeric KlHxk1 in crystal form VII with glucose bound (open state) | | Descriptor: | Hexokinase, SULFATE ION, beta-D-glucopyranose | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-28 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
6HHU
 
 | |
5B6V
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: resting state structure | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nango, E, Royant, A, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
1ZX5
 
 | | The structure of a putative mannosephosphate isomerase from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GLYCEROL, ... | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-06-06 | | Release date: | 2005-07-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a putative mannosephosphate isomerase from Archaeoglobus fulgidus
To be Published
|
|
6HI7
 
 | | The ATAD2 bromodomain in complex with compound 10 | | Descriptor: | (2~{R})-~{N}-[5-(3-aminophenyl)-4-ethanoyl-1,3-thiazol-2-yl]-2-azanyl-propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2018-08-29 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.743 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
