1B43
 
 | | FEN-1 FROM P. FURIOSUS | | Descriptor: | PROTEIN (FEN-1) | | Authors: | Hosfield, D.J, Mol, C.D, Shen, B, Tainer, J.A. | | Deposit date: | 1999-01-05 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity.
Cell(Cambridge,Mass.), 95, 1998
|
|
6CKR
 
 | | Crystal Structure of BRD4 with QC4956 | | Descriptor: | Bromodomain-containing protein 4, N-{3-[2-methyl-6-(1-methyl-1H-pyrazol-4-yl)-1-oxo-1,2-dihydroisoquinolin-4-yl]phenyl}methanesulfonamide | | Authors: | Hosfield, D.J. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Design, synthesis and biological evaluation of novel 4-phenylisoquinolinone BET bromodomain inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6W4K
 
 | | Crystal structure of Lysine Specific Demethylase 1 (LSD1) with CC-90011 | | Descriptor: | 4-[2-(4-aminopiperidin-1-yl)-5-(3-fluoro-4-methoxyphenyl)-1-methyl-6-oxo-1,6-dihydropyrimidin-4-yl]-2-fluorobenzonitrile, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, ... | | Authors: | Hosfield, D.J. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Discovery of CC-90011: A Potent and Selective Reversible Inhibitor of Lysine Specific Demethylase 1 (LSD1).
J.Med.Chem., 63, 2020
|
|
6CG1
 
 | | Crystal Structure of KDM4A with Compound 14 | | Descriptor: | 3-{[(4-fluorophenyl)methyl]amino}pyridine-4-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Hosfield, D.J, Nie, Z. | | Deposit date: | 2018-02-19 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-based design and discovery of potent and selective KDM5 inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6CG2
 
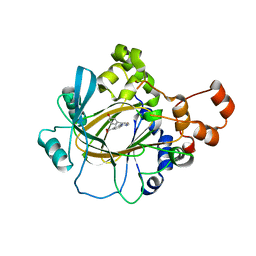 | | Crystal Structure of KDM4A with Compound 8 | | Descriptor: | 2-[5-(3-hydroxyphenyl)-1H-pyrazol-1-yl]pyridine-4-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Hosfield, D.J, Nie, Z. | | Deposit date: | 2018-02-19 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure-based design and discovery of potent and selective KDM5 inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6CKS
 
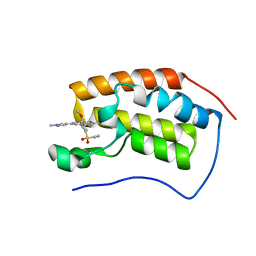 | | Crystal Structure of BRD4 with QC4956 | | Descriptor: | 4-[5-(ethylsulfonyl)-2-methoxyphenyl]-2-methyl-6-(1-methyl-1H-pyrazol-4-yl)isoquinolin-1(2H)-one, Bromodomain-containing protein 4 | | Authors: | Hosfield, D.J. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Design, synthesis and biological evaluation of novel 4-phenylisoquinolinone BET bromodomain inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
7UJY
 
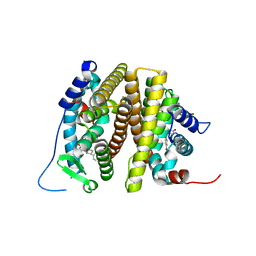 | | Estrogen receptor alpha ligand binding domain Y537S mutant in complex with a methylated lasofoxifene derivative that enhances estrogen receptor alpha nuclear resonance time | | Descriptor: | (5R,6S)-5-(4-{2-[(2R)-2-methylpyrrolidin-1-yl]ethoxy}phenyl)-6-phenyl-5,6,7,8-tetrahydronaphthalen-2-ol, Estrogen receptor | | Authors: | Hosfield, D.J, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-03-31 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Stereospecific lasofoxifene derivatives reveal the interplay between estrogen receptor alpha stability and antagonistic activity in ESR1 mutant breast cancer cells.
Elife, 11, 2022
|
|
7UJF
 
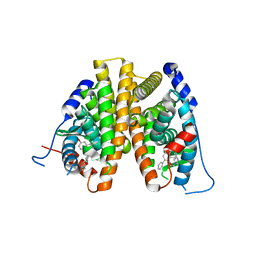 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with a Methylated Lasofoxifene Derivative with Selective Estrogen Receptor Degrader Properties | | Descriptor: | (5R,6S)-5-(4-{2-[(3S)-3-methylpyrrolidin-1-yl]ethoxy}phenyl)-6-phenyl-5,6,7,8-tetrahydronaphthalen-2-ol, Estrogen receptor | | Authors: | Hosfield, D.J, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-03-30 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Stereospecific lasofoxifene derivatives reveal the interplay between estrogen receptor alpha stability and antagonistic activity in ESR1 mutant breast cancer cells.
Elife, 11, 2022
|
|
7UJM
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with a Methylated Lasofoxifene Derivative That Increases Receptor Resonance Time in the Nucleus of Breast Cancer Cells | | Descriptor: | (5R,6S)-5-(4-{2-[(2R)-2-methylpyrrolidin-1-yl]ethoxy}phenyl)-6-phenyl-5,6,7,8-tetrahydronaphthalen-2-ol, Estrogen receptor | | Authors: | Hosfield, D.J, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-03-31 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stereospecific lasofoxifene derivatives reveal the interplay between estrogen receptor alpha stability and antagonistic activity in ESR1 mutant breast cancer cells.
Elife, 11, 2022
|
|
5VGI
 
 | | Crystal Structure of KDM4 with the Small Molecule Inhibitor QC6352 | | Descriptor: | 3-[({(1R)-6-[methyl(phenyl)amino]-1,2,3,4-tetrahydronaphthalen-1-yl}methyl)amino]pyridine-4-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Hosfield, D.J. | | Deposit date: | 2017-04-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Design of KDM4 Inhibitors with Antiproliferative Effects in Cancer Models.
ACS Med Chem Lett, 8, 2017
|
|
5VMP
 
 | | Crystal Structure of Human KDM4 with Small Molecule Inhibitor QC5714 | | Descriptor: | 3-({[(1R)-6-methoxy-1,2,3,4-tetrahydronaphthalen-1-yl]methyl}amino)pyridine-4-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Hosfield, D.J. | | Deposit date: | 2017-04-28 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Design of KDM4 Inhibitors with Antiproliferative Effects in Cancer Models.
ACS Med Chem Lett, 8, 2017
|
|
7UJ8
 
 | |
1RQI
 
 | | Active Conformation of Farnesyl Pyrophosphate Synthase Bound to Isopentyl Pyrophosphate and Dimethylallyl S-Thiolodiphosphate | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, DIPHOSPHATE, Geranyltranstransferase, ... | | Authors: | Hosfield, D.J, Zhang, Y, Dougan, D.R, Brooun, A, Tari, L.W, Swanson, R.V, Finn, J. | | Deposit date: | 2003-12-05 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis
J.Biol.Chem., 279, 2004
|
|
4J52
 
 | | Crystal structure of PLK1 in complex with a pyrimidodiazepinone inhibitor | | Descriptor: | 4-{[(7R)-9-cyclopentyl-7-ethenyl-7-fluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl]amino}-3-methoxy-N-(4-methylpiperazin-1-yl)benzamide, ACETATE ION, Serine/threonine-protein kinase PLK1, ... | | Authors: | Hosfield, D.J, Skene, R.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of TAK-960: An orally available small molecule inhibitor of polo-like kinase 1 (PLK1).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1RQJ
 
 | | Active Conformation of Farnesyl Pyrophosphate Synthase Bound to Isopentyl Pyrophosphate and Risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranyltranstransferase, ... | | Authors: | Hosfield, D.J, Zhang, Y, Dougan, D.R, Brooun, A, Tari, L.W, Swanson, R.V, Finn, J. | | Deposit date: | 2003-12-05 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis
J.Biol.Chem., 279, 2004
|
|
4J53
 
 | | Crystal structure of PLK1 in complex with TAK-960 | | Descriptor: | 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-2-fluoro-5-methoxy-N-(1-methylpiperidin-4-yl)benzamide, ACETATE ION, Serine/threonine-protein kinase PLK1, ... | | Authors: | Hosfield, D.J, Skene, R.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of TAK-960: An orally available small molecule inhibitor of polo-like kinase 1 (PLK1).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1XU7
 
 | | Crystal Structure of the Interface Open Conformation of Tetrameric 11b-HSD1 | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Corticosteroid 11-beta-dehydrogenase, isozyme 1, ... | | Authors: | Hosfield, D.J, Wu, Y, Skene, R.J, Hilger, M, Jennings, A, Snell, G.P, Aertgeerts, K. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational Flexibility in Crystal Structures of Human 11beta-hydroxysteroid dehydrogenase type I provide insights into glucocorticoid interconversion and enzyme regulation.
J.Biol.Chem., 280, 2005
|
|
1XU9
 
 | | Crystal Structure of the Interface Closed Conformation of 11b-hydroxysteroid dehydrogenase isozyme 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Corticosteroid 11-beta-dehydrogenase, ... | | Authors: | Hosfield, D.J, Wu, Y, Skene, R.J, Hilger, M, Jennings, A, Snell, G.P, Aertgeerts, K. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational Flexibility in Crystal Structures of Human 11beta-hydroxysteroid dehydrogenase type I provide insights into glucocorticoid interconversion and enzyme regulation.
J.Biol.Chem., 280, 2005
|
|
7UJO
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with RU39411 | | Descriptor: | (9beta,11beta,17beta)-11-{4-[2-(dimethylamino)ethoxy]phenyl}estra-1,3,5(10)-triene-3,17-diol, Estrogen receptor, MAGNESIUM ION | | Authors: | Hosfield, D.J, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-03-31 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Stereospecific lasofoxifene derivatives reveal the interplay between estrogen receptor alpha stability and antagonistic activity in ESR1 mutant breast cancer cells.
Elife, 11, 2022
|
|
7UJW
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with a Methylated Lasofoxifene Derivative that Possesses Selective Estrogen Receptor Degrader Activities | | Descriptor: | (5R,6S)-5-(4-{2-[(3S)-3-methylpyrrolidin-1-yl]ethoxy}phenyl)-6-phenyl-5,6,7,8-tetrahydronaphthalen-2-ol, Estrogen receptor | | Authors: | Hosfield, D.J, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-03-31 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Stereospecific lasofoxifene derivatives reveal the interplay between estrogen receptor alpha stability and antagonistic activity in ESR1 mutant breast cancer cells.
Elife, 11, 2022
|
|
7UJC
 
 | |
4ISF
 
 | | Human glucokinase in complex with novel activator (2S)-3-cyclohexyl-2-(6-fluoro-2,4-dioxo-1,4-dihydroquinazolin-3(2H)-yl)-N-(1,3-thiazol-2-yl)propanamide | | Descriptor: | (2S)-3-cyclohexyl-2-(6-fluoro-2,4-dioxo-1,4-dihydroquinazolin-3(2H)-yl)-N-(1,3-thiazol-2-yl)propanamide, Glucokinase, IODIDE ION, ... | | Authors: | Hosfield, D, Skene, R.J. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Design, synthesis and SAR of novel glucokinase activators.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4ISE
 
 | | Human glucokinase in complex with novel activator (2S)-3-cyclohexyl-2-(6-fluoro-4-oxoquinazolin-3(4H)-yl)-N-(1,3-thiazol-2-yl)propanamide | | Descriptor: | (2S)-3-cyclohexyl-2-(6-fluoro-4-oxoquinazolin-3(4H)-yl)-N-(1,3-thiazol-2-yl)propanamide, Glucokinase, IODIDE ION, ... | | Authors: | Hosfield, D, Skene, R.J. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Design, synthesis and SAR of novel glucokinase activators.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4ISG
 
 | | Human glucokinase in complex with novel activator (2S)-3-cyclohexyl-2-[4-(methylsulfonyl)-2-oxopiperazin-1-yl]-N-(1,3-thiazol-2-yl)propanamide | | Descriptor: | (2S)-3-cyclohexyl-2-[4-(methylsulfonyl)-2-oxopiperazin-1-yl]-N-(1,3-thiazol-2-yl)propanamide, Glucokinase, IODIDE ION, ... | | Authors: | Hosfield, D, Skene, R.J. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Design, synthesis and SAR of novel glucokinase activators.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1QUM
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI ENDONUCLEASE IV IN COMPLEX WITH DAMAGED DNA | | Descriptor: | 5'-D(*(3DR)P*CP*GP*AP*CP*GP*A)-3', 5'-D(*CP*GP*TP*CP*C)-3', 5'-D(*TP*CP*GP*TP*CP*GP*GP*GP*GP*AP*CP*G)-3', ... | | Authors: | Hosfield, D.J, Guan, Y, Haas, B.J, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 1999-07-01 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis.
Cell(Cambridge,Mass.), 98, 1999
|
|
