2R78
 
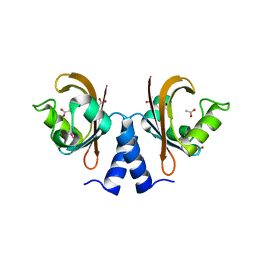 | |
3CK6
 
 | | Crystal structure of ZntB cytoplasmic domain from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | CHLORIDE ION, Putative membrane transport protein | | Authors: | Tan, K, Sather, A, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-14 | | Release date: | 2008-03-25 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and electrostatic property of cytoplasmic domain of ZntB transporter.
Protein Sci., 18, 2009
|
|
3E0X
 
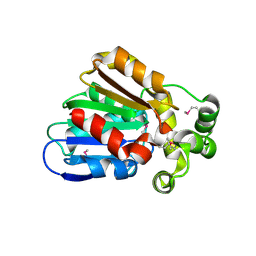 | |
2R4Q
 
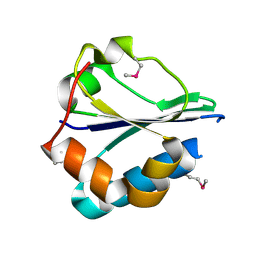 | | The structure of a domain of fruA from Bacillus subtilis | | Descriptor: | Phosphotransferase system (PTS) fructose-specific enzyme IIABC component | | Authors: | Cuff, M.E, Sather, A, Nocek, B, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-31 | | Release date: | 2007-09-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of a domain of fruA from Bacillus subtilis.
TO BE PUBLISHED
|
|
3LW7
 
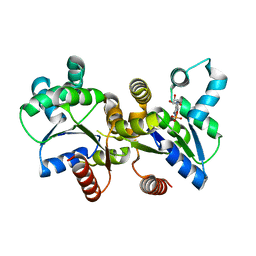 | | The Crystal Structure of an Adenylate kinase-related protein bound to AMP from sulfolobus solfataricus to 2.3A | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenylate kinase related protein (AdkA-like) | | Authors: | Stein, A.J, Sather, A, Hendricks, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-23 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of an Adenylate kinase-related protein bound to AMP from sulfolobus solfataricus to 2.3A
To be Published
|
|
2QMA
 
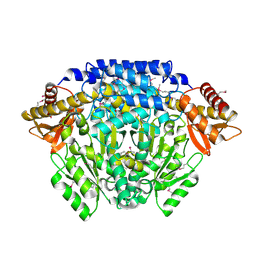 | | Crystal structure of glutamate decarboxylase domain of diaminobutyrate-pyruvate transaminase and L-2,4-diaminobutyrate decarboxylase from Vibrio parahaemolyticus | | Descriptor: | 1,2-ETHANEDIOL, Diaminobutyrate-pyruvate transaminase and L-2,4-diaminobutyrate decarboxylase | | Authors: | Osipiuk, J, Sather, A, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-14 | | Release date: | 2007-07-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray crystal structure of glutamate decarboxylase domain of diaminobutyrate-pyruvate transaminase and L-2,4-diaminobutyrate decarboxylase from Vibrio parahaemolyticus.
To be Published
|
|
2QWV
 
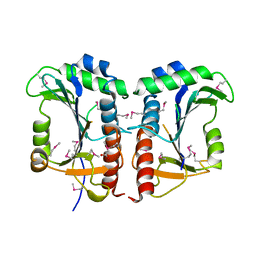 | | Crystal structure of unknown function protein VCA1059 | | Descriptor: | ACETIC ACID, UPF0217 protein VC_A1059 | | Authors: | Chang, C, Sather, A, Moy, S, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-10 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of unknown function protein VCA1059.
To be Published
|
|
3FK8
 
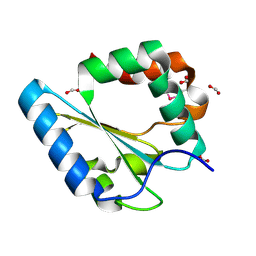 | |
3C7J
 
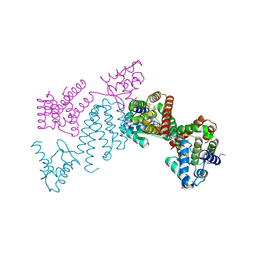 | | Crystal structure of transcriptional regulator (GntR family member) from Pseudomonas syringae pv. tomato str. DC3000 | | Descriptor: | NICKEL (II) ION, Transcriptional regulator, GntR family | | Authors: | Nocek, B, Sather, A, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-07 | | Release date: | 2008-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of transcriptional regulator (GntR family member) from Pseudomonas syringae pv. tomato str. DC3000.
To be Published
|
|
3FEU
 
 | |
2R48
 
 | | Crystal structure of the fructose specific IIB subunit of PTS system from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | Phosphotransferase system (PTS) mannose-specific enzyme IIBCA component | | Authors: | Nocek, B, Cuff, M, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-30 | | Release date: | 2007-09-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the fructose specific IIB subunit of PTS system from Bacillus subtilis subsp. subtilis str. 168.
To be Published
|
|
3LSO
 
 | |
3B33
 
 | |
3G74
 
 | | Crystal structure of a functionally unknown protein from Eubacterium ventriosum ATCC 27560 | | Descriptor: | Protein of unknown function, SULFATE ION | | Authors: | Tan, K, Sather, A, Marshall, N, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-09 | | Release date: | 2009-03-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The crystal structure of a functionally unknown protein from Eubacterium ventriosum ATCC 27560
To be Published
|
|
3G8W
 
 | | Crystal structure of a probable acetyltransferase from Staphylococcus epidermidis ATCC 12228 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CITRATE ANION, Lactococcal prophage ps3 protein 05 | | Authors: | Tan, K, Sather, A, Marshall, N, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of a probable acetyltransferase from Staphylococcus epidermidis ATCC 12228.
To be Published
|
|
3H05
 
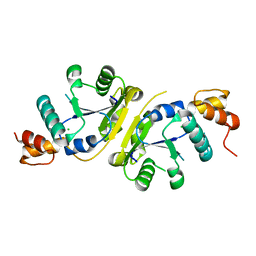 | | The Crystal Structure of a Putative Nicotinate-nucleotide Adenylyltransferase from Vibrio parahaemolyticus | | Descriptor: | CHLORIDE ION, uncharacterized protein VPA0413 | | Authors: | Stein, A.J, Cuff, M.E, Sather, A, Shackelford, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-08 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of a Putative Nicotinate-nucleotide Adenylyltransferase from Vibrio parahaemolyticus
To be Published
|
|
3HDJ
 
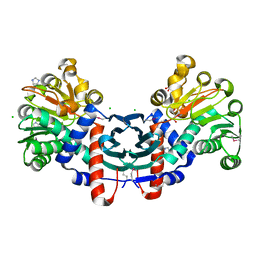 | | The crystal structure of probable ornithine cyclodeaminase from Bordetella pertussis Tohama I | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Tan, K, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-07 | | Release date: | 2009-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of probable ornithine cyclodeaminase from Bordetella pertussis Tohama I
To be Published
|
|
3IDF
 
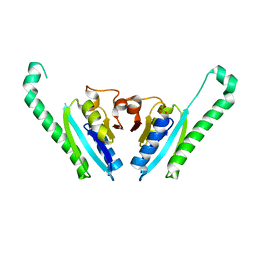 | |
3IS6
 
 | | The Crystal Structure of a domain of a putative Permease protein from Porphyromonas gingivalis to 2A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, putative permease protein, ... | | Authors: | Stein, A.J, Sather, A, Duggan, E, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-25 | | Release date: | 2009-09-08 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of a domain of a putative Permease protein from Porphyromonas gingivalis to 2A
To be Published
|
|
3ICA
 
 | | The crystal structure of the beta subunit of a phenylalanyl-tRNA synthetase from Porphyromonas gingivalis W83 | | Descriptor: | GLYCEROL, Phenylalanyl-tRNA synthetase beta chain, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Fan, Y, Sather, A, Keigher, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-07-28 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The crystal structure of the beta subunit of a phenylalanyl-tRNA synthetase from Porphyromonas gingivalis W83
To be Published
|
|
3IG2
 
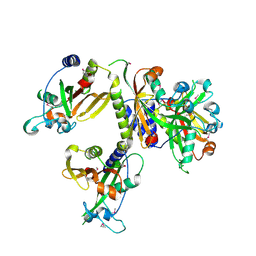 | | The Crystal Structure of a Putative Phenylalanyl-tRNA synthetase (PheRS) beta chain domain from Bacteroides fragilis to 2.1A | | Descriptor: | MAGNESIUM ION, Phenylalanyl-tRNA synthetase beta chain | | Authors: | Stein, A.J, Sather, A, Hendricks, R, Keigher, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-27 | | Release date: | 2009-09-01 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Crystal Structure of a Putative Phenylalanyl-tRNA synthetase (PheRS) beta chain domain from Bacteroides fragilis to 2.1A
To be Published
|
|
3IWF
 
 | | The Crystal Structure of the N-terminal domain of a RpiR Transcriptional Regulator from Staphylococcus epidermidis to 1.4A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-METHOXYETHANOL, CHLORIDE ION, ... | | Authors: | Stein, A.J, Sather, A, Borovilos, M, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-02 | | Release date: | 2009-09-15 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the N-terminal domain of a RpiR Transcriptional Regulator from Staphylococcus epidermidis to 1.4A
To be Published
|
|
3FRM
 
 | | The crystal structure of a functionally unknown conserved protein from Staphylococcus epidermidis ATCC 12228. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SODIUM ION, SULFATE ION, ... | | Authors: | Tan, K, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-08 | | Release date: | 2009-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein from Staphylococcus epidermidis ATCC 12228.
To be Published
|
|
3GBY
 
 | | Crystal structure of a protein with unknown function CT1051 from Chlorobium tepidum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, Uncharacterized protein CT1051 | | Authors: | Fan, Y, Chang, C, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of a protein with unknown function CT1051 from Chlorobium tepidum
To be Published
|
|
3H0K
 
 | |
