1RDK
 
 | | MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH D-GALACTOSE | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-C, ... | | Authors: | Ng, K.K.-S, Drickamer, K, Weis, W.I. | | Deposit date: | 1995-09-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of monosaccharide recognition by rat liver mannose-binding protein.
J.Biol.Chem., 271, 1996
|
|
1RDL
 
 | | MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH ALPHA-METHYL-D-MANNOPYRANOSIDE (0.2 M) | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-C, ... | | Authors: | Ng, K.K.-S, Drickamer, K, Weis, W.I. | | Deposit date: | 1995-09-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of monosaccharide recognition by rat liver mannose-binding protein.
J.Biol.Chem., 271, 1996
|
|
1RDM
 
 | | MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH ALPHA-METHYL-D-MANNOPYRANOSIDE (1.3 M) | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-C, ... | | Authors: | Ng, K.K.-S, Drickamer, K, Weis, W.I. | | Deposit date: | 1995-09-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of monosaccharide recognition by rat liver mannose-binding protein.
J.Biol.Chem., 271, 1996
|
|
1RDO
 
 | | MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-C | | Authors: | Ng, K.K.-S, Drickamer, K, Weis, W.I. | | Deposit date: | 1995-09-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of monosaccharide recognition by rat liver mannose-binding protein.
J.Biol.Chem., 271, 1996
|
|
1RDI
 
 | | MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH ALPHA-METHYL-L-FUCOPYRANOSIDE | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-C, ... | | Authors: | Ng, K.K.-S, Drickamer, K, Weis, W.I. | | Deposit date: | 1995-09-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of monosaccharide recognition by rat liver mannose-binding protein.
J.Biol.Chem., 271, 1996
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.80000925 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
4KZV
 
 | | Structure of the carbohydrate-recognition domain of the C-type lectin mincle bound to trehalose | | Descriptor: | C-type lectin mincle, CALCIUM ION, SODIUM ION, ... | | Authors: | Feinberg, H, Jegouzo, S.A.F, Rowntree, T.J.W, Guan, Y, Brash, M.A, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2013-05-30 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism for Recognition of an Unusual Mycobacterial Glycolipid by the Macrophage Receptor Mincle.
J.Biol.Chem., 288, 2013
|
|
4LDO
 
 | | Structure of beta2 adrenoceptor bound to adrenaline and an engineered nanobody | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, Camelid Antibody Fragment, L-EPINEPHRINE, ... | | Authors: | Ring, A.M, Manglik, A, Kruse, A.C, Enos, M.D, Weis, W.I, Garcia, K.C, Kobilka, B.K. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody.
Nature, 502, 2013
|
|
4LDE
 
 | | Structure of beta2 adrenoceptor bound to BI167107 and an engineered nanobody | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid Antibody Fragment, ... | | Authors: | Ring, A.M, Manglik, A, Kruse, A.C, Enos, M.D, Weis, W.I, Garcia, K.C, Kobilka, B.K. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody.
Nature, 502, 2013
|
|
3KJ6
 
 | | Crystal structure of a Methylated beta2 Adrenergic Receptor-Fab complex | | Descriptor: | Beta-2 adrenergic receptor, Fab heavy chain, Fab light chain, ... | | Authors: | Bokoch, M.P, Zou, Y, Rasmussen, S.G.F, Liu, C.W, Nygaard, R, Rosenbaum, D.M, Fung, J.J, Choi, H.-J, Thian, F.S, Kobilka, T.S, Puglisi, J.D, Weis, W.I, Pardo, L, Prosser, S, Mueller, L, Kobilka, B.K. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor.
Nature, 463, 2010
|
|
3KQG
 
 | | Trimeric Structure of Langerin | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION | | Authors: | Feinberg, H, Powlesland, A.S, Taylor, M.E, Weis, W.I. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trimeric structure of langerin.
J.Biol.Chem., 285, 2010
|
|
1I7X
 
 | | BETA-CATENIN/E-CADHERIN COMPLEX | | Descriptor: | BETA-CATENIN, EPITHELIAL-CADHERIN | | Authors: | Huber, A.H, Weis, W.I. | | Deposit date: | 2001-03-10 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin.
Cell(Cambridge,Mass.), 105, 2001
|
|
1I7W
 
 | | BETA-CATENIN/PHOSPHORYLATED E-CADHERIN COMPLEX | | Descriptor: | BETA-CATENIN, CHLORIDE ION, EPITHELIAL-CADHERIN, ... | | Authors: | Huber, A.H, Weis, W.I. | | Deposit date: | 2001-03-10 | | Release date: | 2001-05-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin.
Cell(Cambridge,Mass.), 105, 2001
|
|
4KMB
 
 | |
5H5M
 
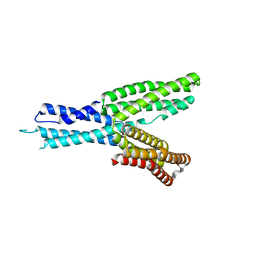 | | Crystal structure of HMP-1 M domain | | Descriptor: | Alpha-catenin-like protein hmp-1 | | Authors: | Kang, H, Bang, I, Weis, W.I, Choi, H.J. | | Deposit date: | 2016-11-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of Caenorhabditis elegans alpha-catenin reveals constitutive binding to beta-catenin and F-actin
J. Biol. Chem., 292, 2017
|
|
2R4R
 
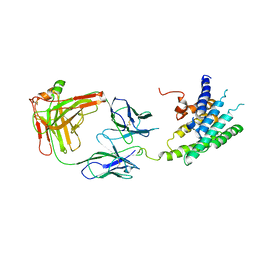 | | Crystal structure of the human beta2 adrenoceptor | | Descriptor: | Beta-2 adrenergic receptor, antibody for beta2 adrenoceptor, heavy chain, ... | | Authors: | Rasmussen, S.G.F, Choi, H.J, Rosenbaum, D.M, Kobilka, T.S, Thian, F.S, Edwards, P.C, Burghammer, M, Ratnala, V.R, Sanishvili, R, Fischetti, R.F, Schertler, G.F, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2007-08-31 | | Release date: | 2007-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the human beta2 adrenergic G-protein-coupled receptor.
Nature, 450, 2007
|
|
8VMG
 
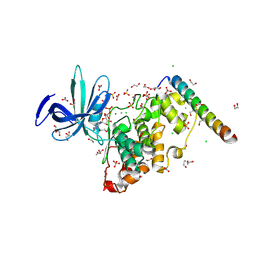 | | Crystal structure of GSK-3 26-383 bound to Axin 383-435 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Enos, M.D, Gavagan, M, Jameson, N, Zalatan, J.G, Weis, W.I. | | Deposit date: | 2024-01-13 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and functional effects of phosphopriming and scaffolding in the kinase GSK-3 beta.
Sci.Signal., 17, 2024
|
|
8VME
 
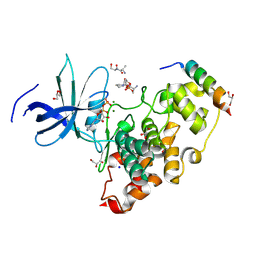 | | Crystal structure of the GSK-3/Axin complex bound to a phosphorylated beta-catenin T41A peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Axin-1, Catenin beta-1, ... | | Authors: | Enos, M.D, Gavagan, M, Jameson, N, Zalatan, J.G, Weis, W.I. | | Deposit date: | 2024-01-13 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional effects of phosphopriming and scaffolding in the kinase GSK-3 beta.
Sci.Signal., 17, 2024
|
|
8VMF
 
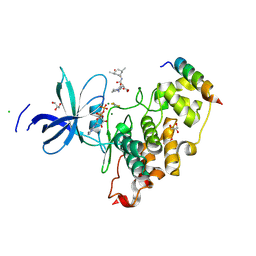 | | Crystal structure of a transition-state mimic of the GSK-3/Axin complex bound to a beta-catenin S45D peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Axin-1, ... | | Authors: | Enos, M.D, Gavagan, M, Jameson, N, Zalatan, J.G, Weis, W.I. | | Deposit date: | 2024-01-13 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional effects of phosphopriming and scaffolding in the kinase GSK-3 beta.
Sci.Signal., 17, 2024
|
|
2BCT
 
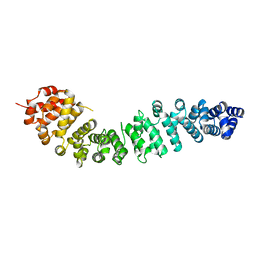 | |
2FO0
 
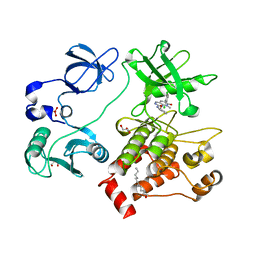 | | Organization of the SH3-SH2 Unit in Active and Inactive Forms of the c-Abl Tyrosine Kinase | | Descriptor: | 6-(2,6-DICHLOROPHENYL)-2-{[3-(HYDROXYMETHYL)PHENYL]AMINO}-8-METHYLPYRIDO[2,3-D]PYRIMIDIN-7(8H)-ONE, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Nagar, B, Hantschel, O, Seeliger, M, Davies, J.M, Weis, W.I, Superti-Furga, G, Kuriyan, J. | | Deposit date: | 2006-01-12 | | Release date: | 2006-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase.
Mol.Cell, 21, 2006
|
|
5JQH
 
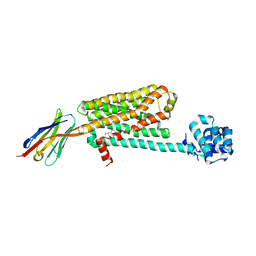 | | Structure of beta2 adrenoceptor bound to carazolol and inactive-state stabilizing nanobody, Nb60 | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, CHOLESTEROL, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Staus, D.P, Strachan, R.T, Manglik, A, Pani, B, Kahsai, A.W, Kim, T.H, Wingler, L.M, Ahn, S, Chatterjee, A, Masoudi, A, Kruse, A.C, Pardon, E, Steyaert, J, Weis, W.I, Prosser, R.S, Kobilka, B.K, Costa, T, Lefkowitz, R.J. | | Deposit date: | 2016-05-05 | | Release date: | 2016-07-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation.
Nature, 535, 2016
|
|
2KMB
 
 | |
3BCT
 
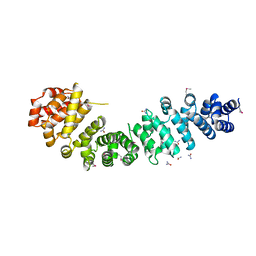 | | THE ARMADILLO REPEAT REGION FROM MURINE BETA-CATENIN | | Descriptor: | BETA-CATENIN, CHLORIDE ION, UREA | | Authors: | Huber, A.H, Nelson, W.J, Weis, W.I. | | Deposit date: | 1997-07-31 | | Release date: | 1997-11-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of the armadillo repeat region of beta-catenin.
Cell(Cambridge,Mass.), 90, 1997
|
|
