3U51
 
 | | Src in complex with DNA-templated macrocyclic inhibitor MC1 | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, macrocyclic inhibitor MC1 | | Authors: | Seeliger, M.A, Liu, D.R, Georghiou, G, Kleiner, R.E, Pulkoski-Gross, M. | | Deposit date: | 2011-10-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | Highly specific, bisubstrate-competitive Src inhibitors from DNA-templated macrocycles.
Nat.Chem.Biol., 8, 2012
|
|
3U4W
 
 | | Src in complex with DNA-templated macrocyclic inhibitor MC4b | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase Src, SULFATE ION, ... | | Authors: | Seeliger, M.A, Liu, D.R, Georghiou, G, Kleiner, R.E, Pulkoski-Gross, M. | | Deposit date: | 2011-10-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly specific, bisubstrate-competitive Src inhibitors from DNA-templated macrocycles.
Nat.Chem.Biol., 8, 2012
|
|
3G6G
 
 | | Equally potent inhibition of c-Src and Abl by compounds that recognize inactive kinase conformations | | Descriptor: | GLYCEROL, N-{3-[(3-{4-[(4-methoxyphenyl)amino]-1,3,5-triazin-2-yl}pyridin-2-yl)amino]-4-methylphenyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Seeliger, M.A, Ranjitkar, P, Kasap, C, Shan, Y, Shaw, D.E, Shah, N.P, Kuriyan, J, Maly, D.J. | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Equally potent inhibition of c-Src and Abl by compounds that recognize inactive kinase conformations
Cancer Res., 69, 2009
|
|
3G6H
 
 | | Src Thr338Ile inhibited in the DFG-Asp-Out conformation | | Descriptor: | N-{4-methyl-3-[(3-{4-[(3,4,5-trimethoxyphenyl)amino]-1,3,5-triazin-2-yl}pyridin-2-yl)amino]phenyl}-3-(trifluoromethyl)benzamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Seeliger, M.A, Ranjitkar, P, Kasap, C, Shan, Y, Shaw, D.E, Shah, N.P, Kuriyan, J, Maly, D.J. | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Equally potent inhibition of c-Src and Abl by compounds that recognize inactive kinase conformations
Cancer Res., 69, 2009
|
|
3F6X
 
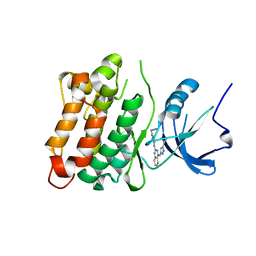 | | c-Src kinase domain in complex with small molecule inhibitor | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, [4-({4-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]quinazolin-2-yl}amino)phenyl]acetonitrile | | Authors: | Seeliger, M.A, Statsuk, A.V, Maly, D.J, Patrick, P.Z, Kuriyan, J, Shokat, K.M. | | Deposit date: | 2008-11-06 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Tuning a three-component reaction for trapping kinase substrate complexes.
J.Am.Chem.Soc., 130, 2008
|
|
2OIQ
 
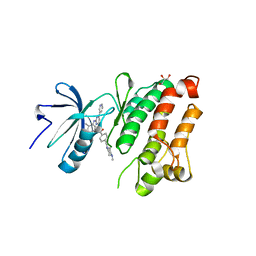 | | Crystal Structure of chicken c-Src kinase domain in complex with the cancer drug imatinib. | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, GLYCEROL, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Seeliger, M.A, Nagar, B, Frank, F, Cao, X, Henderson, M.N, Kuriyan, J. | | Deposit date: | 2007-01-11 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | c-Src Binds to the Cancer Drug Imatinib with an Inactive Abl/c-Kit Conformation and a Distributed Thermodynamic Penalty.
Structure, 15, 2007
|
|
8FE9
 
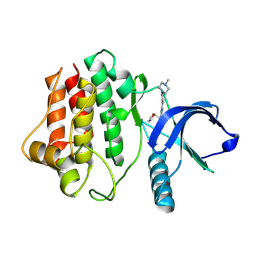 | | Crystal structure of Ack1 kinase K161Q mutant in complex with the selective inhibitor (R)-9b | | Descriptor: | Activated CDC42 kinase 1, N-[(1S)-1-benzyl-2-[(6-chloro-2-oxo-1H-quinolin-4-yl)methylamino]-2-oxo-ethyl]-4-hydroxy-2-oxo-1H-quinoline-6-carbo | | Authors: | Paung, Y, Kan, Y, Seeliger, M.S, Miller, W.T. | | Deposit date: | 2022-12-06 | | Release date: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical Studies of Systemic Lupus Erythematosus-Associated Mutations in Nonreceptor Tyrosine Kinases Ack1 and Brk.
Biochemistry, 62, 2023
|
|
7N99
 
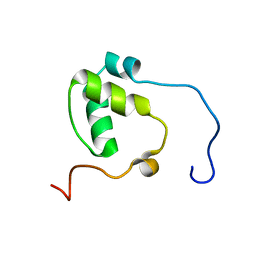 | | SDE2 SAP domain apo structure | | Descriptor: | Isoform 2 of Replication stress response regulator SDE2 | | Authors: | Paung, Y, Weinheimer, A.S, Rageul, J, Khan, A, Ho, B, Tong, M, Alphonse, S, Seeliger, M.A, Kim, H. | | Deposit date: | 2021-06-17 | | Release date: | 2022-10-12 | | Last modified: | 2023-04-26 | | Method: | SOLUTION NMR | | Cite: | Extended DNA-binding interfaces beyond the canonical SAP domain contribute to the function of replication stress regulator SDE2 at DNA replication forks.
J.Biol.Chem., 298, 2022
|
|
6EDS
 
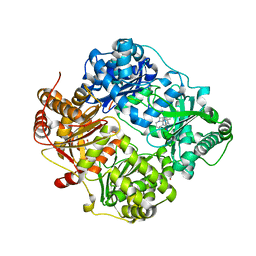 | | Structure of Cysteine-free Human Insulin-Degrading Enzyme in complex with Glucagon and Substrate-selective Macrocyclic Inhibitor 63 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, G.A, Seeliger, M.A, Maianti, J.P, Liu, D.R, Welsh, A.J. | | Deposit date: | 2018-08-10 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.18071723 Å) | | Cite: | Substrate-selective inhibitors that reprogram the activity of insulin-degrading enzyme.
Nat.Chem.Biol., 15, 2019
|
|
6MQ3
 
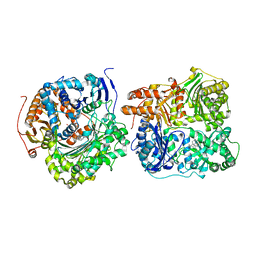 | | Structure of Cysteine-free Human Insulin-Degrading Enzyme in complex with Substrate-selective Macrocycle Inhibitor 63 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Insulin-degrading enzyme, {(8R,9S,10S)-9-(2',3'-dimethyl[1,1'-biphenyl]-4-yl)-6-[(1-methyl-1H-imidazol-2-yl)sulfonyl]-1,6-diazabicyclo[6.2.0]decan-10-yl}methanol | | Authors: | Tan, G.A, Seeliger, M.A, Welsh, A.J, Maianti, J.P, Liu, D.R. | | Deposit date: | 2018-10-09 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.569147 Å) | | Cite: | Substrate-selective inhibitors that reprogram the activity of insulin-degrading enzyme.
Nat.Chem.Biol., 15, 2019
|
|
7KP6
 
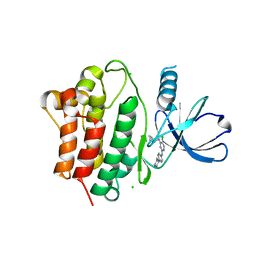 | | Structure of Ack1 kinase in complex with a selective inhibitor | | Descriptor: | 5-chloro-N~2~-[4-(4-methylpiperazin-1-yl)phenyl]-N~4~-{[(2R)-oxolan-2-yl]methyl}pyrimidine-2,4-diamine, Activated CDC42 kinase 1, CHLORIDE ION | | Authors: | Thakur, M.K, Miller, W.T, Mahajan, N, Seeliger, M.A. | | Deposit date: | 2020-11-10 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Inhibiting ACK1-mediated phosphorylation of C-terminal Src kinase counteracts prostate cancer immune checkpoint blockade resistance.
Nat Commun, 13, 2022
|
|
4LTE
 
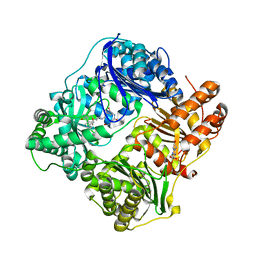 | | Structure of Cysteine-free Human Insulin Degrading Enzyme in Complex with Macrocyclic Inhibitor | | Descriptor: | 2,6-DIAMINO-HEXANOIC ACID AMIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FUMARIC ACID, ... | | Authors: | Foda, Z.H, Seeliger, M.A, Saghatelian, A, Liu, D.R. | | Deposit date: | 2013-07-23 | | Release date: | 2014-05-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Anti-diabetic activity of insulin-degrading enzyme inhibitors mediated by multiple hormones.
Nature, 511, 2014
|
|
5BMM
 
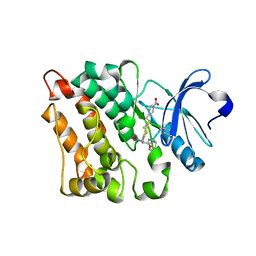 | | Src in complex with DNA-templated macrocyclic inhibitor MC25b | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, macrocyclic inhibitor MC25b | | Authors: | Georghiou, G, Guja, K.E, Aleem, S, Kleiner, R.E, Liu, D.R, Miller, W.T, Garcia-Diaz, M, Seeliger, M.A. | | Deposit date: | 2015-05-22 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Basis for Intracellular Kinase Inhibition by Src-specific Peptidic Macrocycles.
Cell Chem Biol, 23, 2016
|
|
4AGW
 
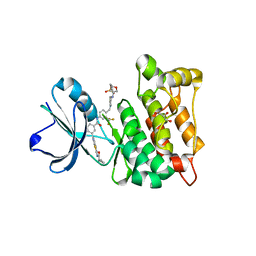 | | Discovery of a small molecule type II inhibitor of wild-type and gatekeeper mutants of BCR-ABL, PDGFRalpha, Kit, and Src kinases | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-{2-[(cyclopropylcarbonyl)amino][1,3]thiazolo[5,4-b]pyridin-5-yl}-N-{4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromet hyl)phenyl}benzamide, GLYCEROL, ... | | Authors: | Weisberg, E, Choi, H.G, Seeliger, M, Gray, N, Griffin, J.D. | | Deposit date: | 2012-02-01 | | Release date: | 2012-02-15 | | Last modified: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a Small-Molecule Type II Inhibitor of Wild-Type and Gatekeeper Mutants of Bcr-Abl, Pdgfralpha, Kit, and Src Kinases: Novel Type II Inhibitor of Gatekeeper Mutants.
Blood, 115, 2010
|
|
2FO0
 
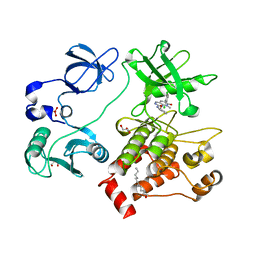 | | Organization of the SH3-SH2 Unit in Active and Inactive Forms of the c-Abl Tyrosine Kinase | | Descriptor: | 6-(2,6-DICHLOROPHENYL)-2-{[3-(HYDROXYMETHYL)PHENYL]AMINO}-8-METHYLPYRIDO[2,3-D]PYRIMIDIN-7(8H)-ONE, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Nagar, B, Hantschel, O, Seeliger, M, Davies, J.M, Weis, W.I, Superti-Furga, G, Kuriyan, J. | | Deposit date: | 2006-01-12 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase.
Mol.Cell, 21, 2006
|
|
7THC
 
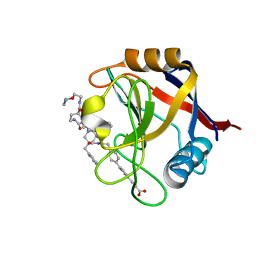 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B25 | | Descriptor: | 3-(4'-{[(4S,7S,11R,13E,19S)-19-{[2-(2-aminoethoxy)ethyl]carbamoyl}-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosin-4-yl]methyl}[1,1'-biphenyl]-4-yl)prop-2-ynoic acid, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7TH1
 
 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B3 | | Descriptor: | (4S,7S,11R,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-7-benzyl-4-[(3-methylphenyl)methyl]-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7TGU
 
 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B1 | | Descriptor: | (4S,7S,11R,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-4-[(4-benzoylphenyl)methyl]-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7TH6
 
 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B21 | | Descriptor: | 4'-{[(4S,7S,11R,13E,19S)-19-{[2-(2-aminoethoxy)ethyl]carbamoyl}-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosin-4-yl]methyl}-2-methyl[1,1'-biphenyl]-4-carboxylic acid, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7THF
 
 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B53 | | Descriptor: | 3-(4'-{[(4S,7S,11R,13E,19S)-19-{[2-(2-aminoethoxy)ethyl]carbamoyl}-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosin-4-yl]methyl}[1,1'-biphenyl]-4-yl)pentanedioic acid, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7TGV
 
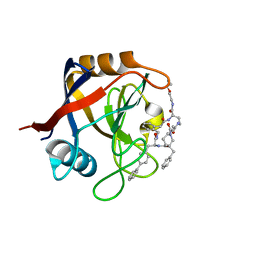 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B2 | | Descriptor: | (4S,7S,11R,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-7-benzyl-4-[([1,1'-biphenyl]-4-yl)methyl]-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7THD
 
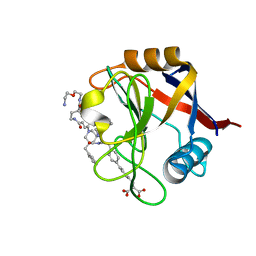 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B52 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F, mitochondrial, [(4'-{[(4S,7S,11R,13E,19S)-19-{[2-(2-aminoethoxy)ethyl]carbamoyl}-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosin-4-yl]methyl}[1,1'-biphenyl]-4-yl)methyl]propanedioic acid | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7TH7
 
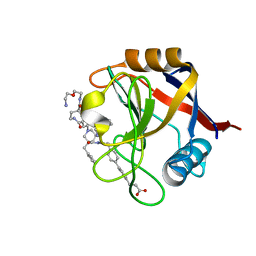 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B23 | | Descriptor: | 3-(4'-{[(4S,7S,11R,19S)-19-{[2-(2-aminoethoxy)ethyl]carbamoyl}-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,13,14,15,16,17,18,19,20,21,22-icosahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosin-4-yl]methyl}[1,1'-biphenyl]-4-yl)propanoic acid, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7TGS
 
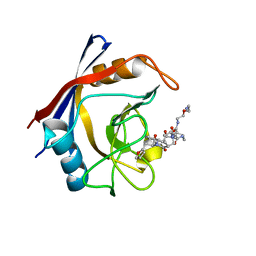 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor JOMBt | | Descriptor: | (4S,7R,11S,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-4-[(furan-2-yl)methyl]-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7TGT
 
 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor A26 | | Descriptor: | (4S,7S,11R,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-7-benzyl-4-[(furan-2-yl)methyl]-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
