8S7J
 
 | | Coxsackievirus A9 bound with compound 20 (CL300) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Plavec, Z, Butcher, S.J, Mitchell, C, Buckner, C. | | Deposit date: | 2024-03-01 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | SAR Analysis of Novel Coxsackie virus A9 Capsid Binders.
J.Med.Chem., 67, 2024
|
|
5IXD
 
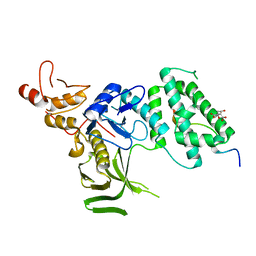 | |
4N7M
 
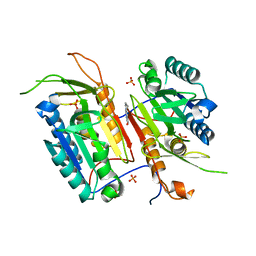 | |
4LXZ
 
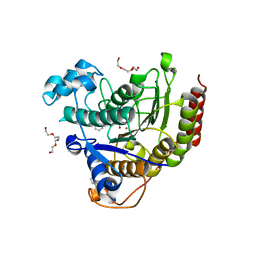 | | Structure of Human HDAC2 in complex with SAHA (vorinostat) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, Histone deacetylase 2, ... | | Authors: | Fong, R, Lupardus, P.J. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Histone Deacetylase (HDAC) Inhibitor Kinetic Rate Constants Correlate with Cellular Histone Acetylation but Not Transcription and Cell Viability.
J.Biol.Chem., 288, 2013
|
|
2M0X
 
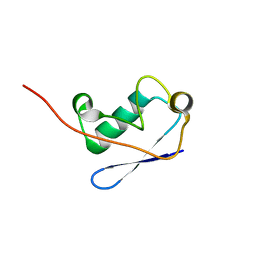 | |
4RJ3
 
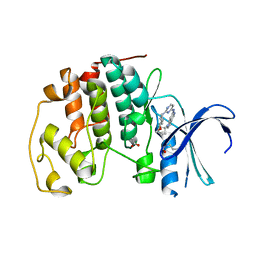 | | CDK2 with EGFR inhibitor compound 8 | | Descriptor: | 1-cyclopentyl-N-[2-(4-methoxypiperidin-1-yl)pyrimidin-4-yl]-1H-pyrrolo[3,2-c]pyridin-6-amine, ACETATE ION, Cyclin-dependent kinase 2 | | Authors: | Eigenbrot, C, Yin, J. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-26 | | Last modified: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of Selective and Noncovalent Diaminopyrimidine-Based Inhibitors of Epidermal Growth Factor Receptor Containing the T790M Resistance Mutation.
J.Med.Chem., 57, 2014
|
|
4RJ4
 
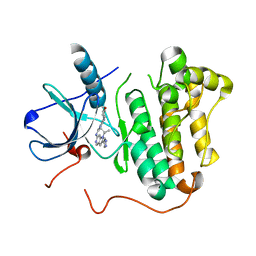 | | EGFR kinase (T790M/L858R) with inhibitor compound 6 | | Descriptor: | Epidermal growth factor receptor, N-[2-(4-methoxypiperidin-1-yl)pyrimidin-4-yl]-1-(propan-2-yl)-2-(1H-pyrazol-4-yl)-1H-pyrrolo[3,2-c]pyridin-6-amine | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Discovery of Selective and Noncovalent Diaminopyrimidine-Based Inhibitors of Epidermal Growth Factor Receptor Containing the T790M Resistance Mutation.
J.Med.Chem., 57, 2014
|
|
4RJ6
 
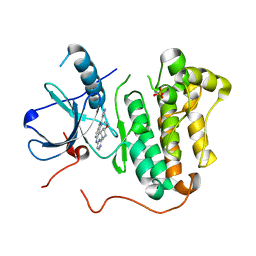 | | EGFR kinase (T790M/L858R) with inhibitor compound 4 | | Descriptor: | Epidermal growth factor receptor, N-[2-(4-methoxypiperidin-1-yl)pyrimidin-4-yl]-2-(1H-pyrazol-4-yl)-3H-imidazo[4,5-c]pyridin-6-amine, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Selective and Noncovalent Diaminopyrimidine-Based Inhibitors of Epidermal Growth Factor Receptor Containing the T790M Resistance Mutation.
J.Med.Chem., 57, 2014
|
|
4RJ8
 
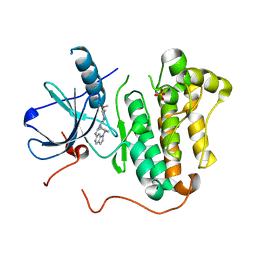 | | EGFR kinase (T790M/L858R) with inhibitor compound 8 | | Descriptor: | 1-cyclopentyl-N-[2-(4-methoxypiperidin-1-yl)pyrimidin-4-yl]-1H-pyrrolo[3,2-c]pyridin-6-amine, Epidermal growth factor receptor, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Selective and Noncovalent Diaminopyrimidine-Based Inhibitors of Epidermal Growth Factor Receptor Containing the T790M Resistance Mutation.
J.Med.Chem., 57, 2014
|
|
4RJ5
 
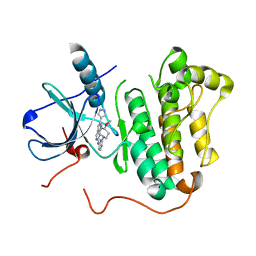 | | EGFR kinase (T790M/L858R) with inhibitor compound 5 | | Descriptor: | Epidermal growth factor receptor, N-[2-(4-methoxypiperidin-1-yl)pyrimidin-4-yl]-2-(1H-pyrazol-4-yl)-1H-pyrrolo[3,2-c]pyridin-6-amine | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of Selective and Noncovalent Diaminopyrimidine-Based Inhibitors of Epidermal Growth Factor Receptor Containing the T790M Resistance Mutation.
J.Med.Chem., 57, 2014
|
|
4RJ7
 
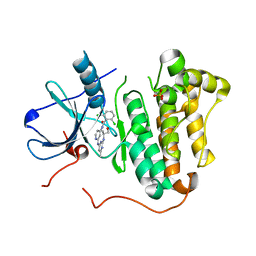 | | EGFR kinase (T790M/L858R) with inhibitor compound 1 | | Descriptor: | 2,6-dichloro-N-{2-[(2-{[(2S)-1-hydroxypropan-2-yl]amino}-6-methylpyrimidin-4-yl)amino]pyridin-4-yl}benzamide, Epidermal growth factor receptor, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of Selective and Noncovalent Diaminopyrimidine-Based Inhibitors of Epidermal Growth Factor Receptor Containing the T790M Resistance Mutation.
J.Med.Chem., 57, 2014
|
|
5KI0
 
 | | NMR structure of human antimicrobial peptide KAMP-19 | | Descriptor: | Antimicrobial peptide KAMP-19 | | Authors: | Wang, G. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Membrane-Active Epithelial Keratin 6A Fragments (KAMPs) Are Unique Human Antimicrobial Peptides with a Non-alpha beta Structure.
Front Microbiol, 7, 2016
|
|
6WJ5
 
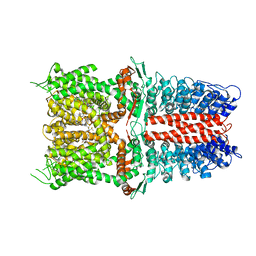 | | Structure of human TRPA1 in complex with inhibitor GDC-0334 | | Descriptor: | (4R,5S)-4-fluoro-1-[(4-fluorophenyl)sulfonyl]-5-methyl-N-({5-(trifluoromethyl)-2-[2-(trifluoromethyl)pyrimidin-5-yl]pyridin-4-yl}methyl)-L-prolinamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Arthur, C.P, Volgraf, M, Chen, H. | | Deposit date: | 2020-04-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A TRPA1 inhibitor suppresses neurogenic inflammation and airway contraction for asthma treatment.
J.Exp.Med., 218, 2021
|
|
5KUP
 
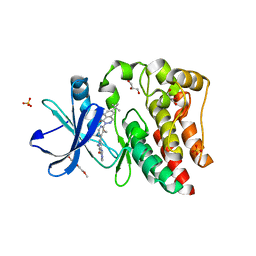 | | Bruton's tyrosine kinase (BTK) with pyridazinone compound 9 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 6-~{tert}-butyl-8-fluoranyl-2-[3-(hydroxymethyl)-4-[1-methyl-6-oxidanylidene-5-(pyrimidin-4-ylamino)pyridin-3-yl]pyridin-2-yl]phthalazin-1-one, GLYCEROL, ... | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.389 Å) | | Cite: | Battling Btk Mutants With Noncovalent Inhibitors That Overcome Cys481 and Thr474 Mutations.
Acs Chem.Biol., 11, 2016
|
|
5IXI
 
 | |
7TJ8
 
 | | Cryo-EM structure of the human Nax channel in complex with beta3 solved in nanodiscs | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Noland, C.L, Kschonsak, M, Ciferri, C, Payandeh, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-guided unlocking of Na X reveals a non-selective tetrodotoxin-sensitive cation channel.
Nat Commun, 13, 2022
|
|
7TJ9
 
 | | Cryo-EM structure of the human Nax channel in complex with beta3 solved in GDN | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Noland, C.L, Kschonsak, M, Ciferri, C, Payandeh, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-guided unlocking of Na X reveals a non-selective tetrodotoxin-sensitive cation channel.
Nat Commun, 13, 2022
|
|
5EK0
 
 | | Human Nav1.7-VSD4-NavAb in complex with GX-936. | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-cyano-4-[2-[2-(1-ethylazetidin-3-yl)pyrazol-3-yl]-4-(trifluoromethyl)phenoxy]-~{N}-(1,2,4-thiadiazol-5-yl)benzenesulfonamide, Chimera of bacterial Ion transport protein and human Sodium channel protein type 9 subunit alpha | | Authors: | Ahuja, S, Mukund, S, Starovasnik, M.A, Koth, C.M, Payandeh, J. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist.
Science, 350, 2015
|
|
8TLM
 
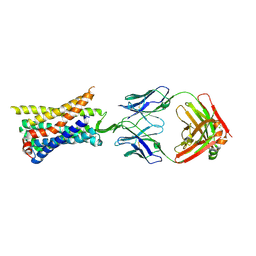 | | Structure of a class A GPCR/Fab complex | | Descriptor: | C-C chemokine receptor type 8, Green fluorescent protein fusion, Fab heavy chain, ... | | Authors: | Sun, D, Johnson, M, Masureel, M. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
8U1U
 
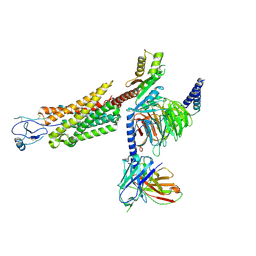 | | Structure of a class A GPCR/agonist complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-C motif chemokine 1,C-C chemokine receptor type 8,EGFP fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Sun, D, Johnson, M, Masureel, M. | | Deposit date: | 2023-09-02 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
7UJD
 
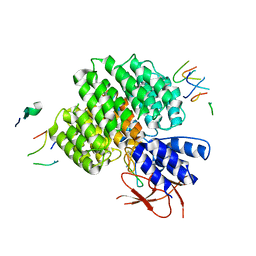 | | PSMD2 Structure bound to MC1 and Fab8/14 | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 2, ACY-PHE-PRO-ASP-VAL-SAR-LEU-HIS-ARG-TYR-TRP-GLY-TRP-ASP-CYS-GLY-NH2, Fab 14 HC CDRs, ... | | Authors: | Johnson, M.C, Bashore, C, Ciferri, C, Dueber, E.C. | | Deposit date: | 2022-03-30 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Targeted degradation via direct 26S proteasome recruitment.
Nat.Chem.Biol., 19, 2023
|
|
7UIH
 
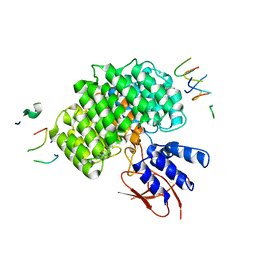 | | PSMD2 Structure | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 2, Fab 14 HC CDRs, Fab 14 LC CDRs, ... | | Authors: | Johnson, M.C, Bashore, C, Ciferri, C, Dueber, E.C. | | Deposit date: | 2022-03-29 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Targeted degradation via direct 26S proteasome recruitment.
Nat.Chem.Biol., 19, 2023
|
|
7RP3
 
 | | Crystal structure of GNE-1952 alkylated KRAS G12C in complex with 2H11 CLAMP | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[6-chloranyl-7-(5-methyl-1~{H}-indazol-4-yl)quinazolin-4-yl]piperazin-1-yl]propan-1-one, GLYCEROL, ... | | Authors: | Oh, A, Tam, C, Wang, W. | | Deposit date: | 2021-08-03 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformation-locking antibodies for the discovery and characterization of KRAS inhibitors.
Nat.Biotechnol., 40, 2022
|
|
7RP2
 
 | | Crystal structure of Kas G12C in complex with 2H11 CLAMP | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, GTPase KRas, ... | | Authors: | Oh, A, Tam, C, Wang, W. | | Deposit date: | 2021-08-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformation-locking antibodies for the discovery and characterization of KRAS inhibitors.
Nat.Biotechnol., 40, 2022
|
|
7RP4
 
 | | Crystal structure of KRAS G12C in complex with GNE-1952 | | Descriptor: | 1-[4-[6-chloranyl-7-(5-methyl-1~{H}-indazol-4-yl)quinazolin-4-yl]piperazin-1-yl]propan-1-one, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Oh, A, Tam, C, Wang, W. | | Deposit date: | 2021-08-03 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformation-locking antibodies for the discovery and characterization of KRAS inhibitors.
Nat.Biotechnol., 40, 2022
|
|
