4C9Y
 
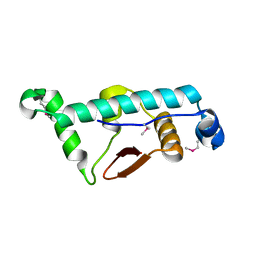 | | Structural Basis for the microtubule binding of the human kinetochore Ska complex | | Descriptor: | SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 1 | | Authors: | Abad, M, Medina, B, Santamaria, A, Zou, J, Plasberg-Hill, C, Madhumalar, A, Jayachandran, U, Redli, P.M, Rappsilber, J, Nigg, E.A, Jeyaprakash, A.A. | | Deposit date: | 2013-10-04 | | Release date: | 2014-01-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis for Microtubule Recognition by the Human Kinetochore Ska Complex.
Nat.Commun., 5, 2014
|
|
8CKB
 
 | | Asymmetric reconstruction of the crAss001 virion | | Descriptor: | Auxiliary capsid protein gp36, Cargo protein 1 gp45, Head fiber dimer protein gp29, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2023-02-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.39 Å) | | Cite: | Structural atlas of the most abundant human gut virus
Nature
|
|
2L9X
 
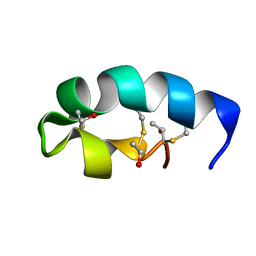 | | Trn- peptide of the two-component bacteriocin Thuricin CD | | Descriptor: | Uncharacterized protein | | Authors: | Sit, C.S, Mckay, R.T, Hill, C, Ross, R.P, Vederas, J.C. | | Deposit date: | 2011-02-25 | | Release date: | 2012-01-11 | | Last modified: | 2018-08-22 | | Method: | SOLUTION NMR | | Cite: | The 3D structure of thuricin CD, a two-component bacteriocin with cysteine sulfur to alpha-carbon cross-links.
J.Am.Chem.Soc., 133, 2011
|
|
2LA0
 
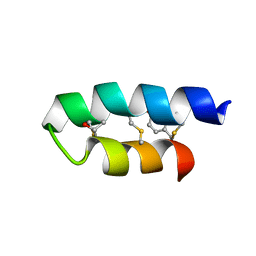 | | Trn- peptide of the two-component bacteriocin Thuricin CD | | Descriptor: | Uncharacterized protein | | Authors: | Sit, C.S, Mckay, R.T, Hill, C, Ross, R.P, Vederas, J.C. | | Deposit date: | 2011-02-27 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The 3D structure of thuricin CD, a two-component bacteriocin with cysteine sulfur to alpha-carbon cross-links.
J.Am.Chem.Soc., 133, 2011
|
|
2OPG
 
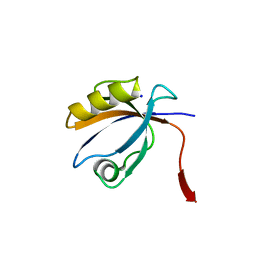 | | The crystal structure of the 10th PDZ domain of MPDZ | | Descriptor: | Multiple PDZ domain protein, SODIUM ION | | Authors: | Gileadi, C, Phillips, C, Elkins, J, Papagrigoriou, E, Ugochukwu, E, Gorrec, F, Savitsky, P, Umeano, C, Berridge, G, Gileadi, O, Salah, E, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the 10th PDZ domain of MPDZ
To be Published
|
|
4V01
 
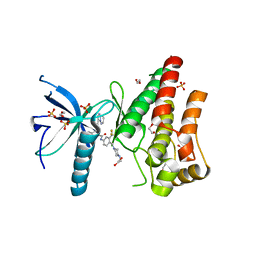 | | FGFR1 in complex with ponatinib (co-crystallisation). | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, ... | | Authors: | Tucker, J, Klein, T, Breed, J, Breeze, A, Overman, R, Phillips, C, Norman, R.A. | | Deposit date: | 2014-09-10 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural Insights Into Fgfr Kinase Isoform Selectivity: Diverse Binding Modes of Azd4547 and Ponatinib in Complex with Fgfr1 and Fgfr4
Structure, 22, 2014
|
|
4V05
 
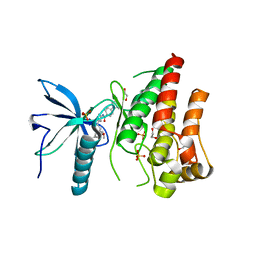 | | FGFR1 in complex with AZD4547. | | Descriptor: | 1,2-ETHANEDIOL, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, PFEIFFER SYNDROME), ... | | Authors: | Tucker, J, Klein, T, Breed, J, Breeze, A, Overman, R, Phillips, C, Norman, R.A. | | Deposit date: | 2014-09-10 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural Insights Into Fgfr Kinase Isoform Selectivity: Diverse Binding Modes of Azd4547 and Ponatinib in Complex with Fgfr1 and Fgfr4
Structure, 22, 2014
|
|
3NR3
 
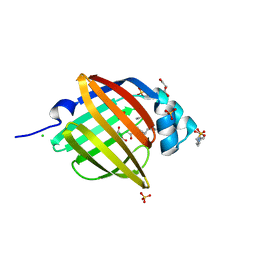 | | Crystal Structure of Human Peripheral Myelin Protein 2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Ugochukwu, E, Pilka, E, Phillips, C, Yue, W.W, Krojer, T, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Human Peripheral Myelin Protein 2
To be Published
|
|
4UY9
 
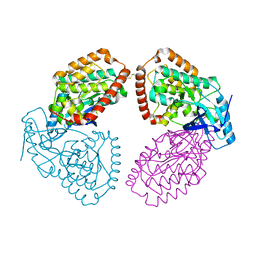 | | Structure of MLK1 kinase domain with leucine zipper 1 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 9 | | Authors: | Read, J.A, Brassington, C, Pollard, H.K, Phillips, C, Green, I, Overmann, R, Collier, M. | | Deposit date: | 2014-08-29 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Recurrent Mlk4 Loss-of-Function Mutations Suppress Jnk Signaling to Promote Colon Tumorigenesis.
Cancer Res., 76, 2016
|
|
4UYA
 
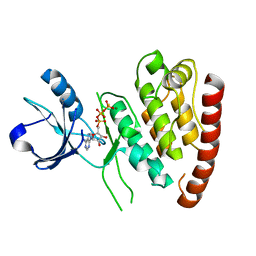 | | Structure of MLK4 kinase domain with ATPgammaS | | Descriptor: | MAGNESIUM ION, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE MLK4, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Read, J.A, Brassington, C, Pollard, H.K, Phillips, C, Green, I, Overmann, R, Collier, M. | | Deposit date: | 2014-08-29 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recurrent Mlk4 Loss-of-Function Mutations Suppress Jnk Signaling to Promote Colon Tumorigenesis.
Cancer Res., 76, 2016
|
|
4V04
 
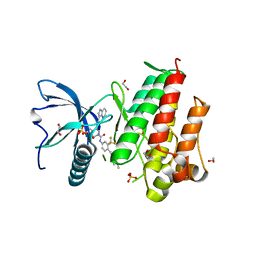 | | FGFR1 in complex with ponatinib. | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, ... | | Authors: | Tucker, J, Klein, T, Breed, J, Breeze, A, Overman, R, Phillips, C, Norman, R.A. | | Deposit date: | 2014-09-10 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Insights Into Fgfr Kinase Isoform Selectivity: Diverse Binding Modes of Azd4547 and Ponatinib in Complex with Fgfr1 and Fgfr4
Structure, 22, 2014
|
|
4UXQ
 
 | | FGFR4 in complex with Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, FIBROBLAST GROWTH FACTOR RECEPTOR 4, SULFATE ION | | Authors: | Tucker, J, Klein, T, Breed, J, Breeze, A, Overman, R, Phillips, C, Norman, R.A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights Into Fgfr Kinase Isoform Selectivity: Diverse Binding Modes of Azd4547 and Ponatinib in Complex with Fgfr1 and Fgfr4
Structure, 22, 2014
|
|
5A4C
 
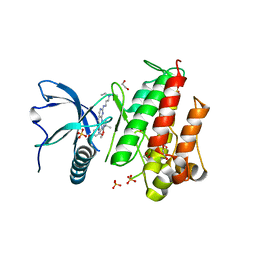 | | FGFR1 ligand complex | | Descriptor: | 1,2-ETHANEDIOL, 1-tert-butyl-3-[2-[3-(diethylamino)propylamino]-6-(3,5-dimethoxyphenyl)pyrido[2,3-d]pyrimidin-7-yl]urea, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, ... | | Authors: | Klein, T, Vajpai, N, Phillips, J.J, Davies, G, Holdgate, G.A, Phillips, C, Tucker, J.A, Norman, R.A, Scott, A.S, Higazi, D.R, Lowe, D, Thompson, G.S, Breeze, A.L. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Dynamic Insights Into the Energetics of Activation Loop Rearrangement in Fgfr1 Kinase.
Nat.Commun., 6, 2015
|
|
5A46
 
 | | FGFR1 in complex with dovitinib | | Descriptor: | 1,2-ETHANEDIOL, 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, ... | | Authors: | Klein, T, Vajpai, N, Phillips, J.J, Davies, G, Holdgate, G.A, Phillips, C, Tucker, J.A, Norman, R.A, Scott, A.S, Higazi, D.R, Lowe, D, Thompson, G.S, Breeze, A.L. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural and Dynamic Insights Into the Energetics of Activation Loop Rearrangement in Fgfr1 Kinase.
Nat.Commun., 6, 2015
|
|
6H47
 
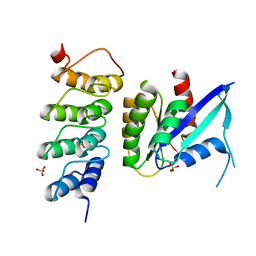 | | Human KRAS in complex with darpin K19 | | Descriptor: | GTPase KRas, SULFATE ION, darpin K19 | | Authors: | Debreczeni, J.E, Bery, N, Legg, S, Breed, J, Embrey, K, Stubbs, C, Kolasinska-Zwierz, P, Barrett, N, Marwood, R, Watson, J, Tart, J, Overman, R, Miller, A, Phillips, C, Minter, R, Rabbitts, T.H. | | Deposit date: | 2018-07-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | KRAS-specific inhibition using a DARPin binding to a site in the allosteric lobe.
Nat Commun, 10, 2019
|
|
1PGO
 
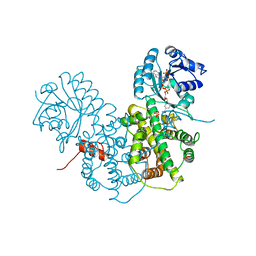 | | CRYSTALLOGRAPHIC STUDY OF COENZYME, COENZYME ANALOGUE AND SUBSTRATE BINDING IN 6-PHOSPHOGLUCONATE DEHYDROGENASE: IMPLICATIONS FOR NADP SPECIFICITY AND THE ENZYME MECHANISM | | Descriptor: | 6-PHOSPHOGLUCONATE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Adams, M.J, Phillips, C, Gover, S. | | Deposit date: | 1994-07-18 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism.
Structure, 2, 1994
|
|
1PGP
 
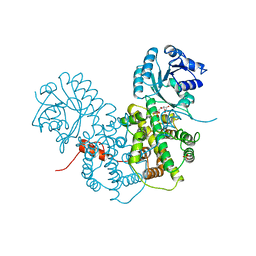 | | CRYSTALLOGRAPHIC STUDY OF COENZYME, COENZYME ANALOGUE AND SUBSTRATE BINDING IN 6-PHOSPHOGLUCONATE DEHYDROGENASE: IMPLICATIONS FOR NADP SPECIFICITY AND THE ENZYME MECHANISM | | Descriptor: | 6-PHOSPHOGLUCONATE DEHYDROGENASE, 6-PHOSPHOGLUCONIC ACID, SULFATE ION | | Authors: | Adams, M.J, Phillips, C, Gover, S, Naylor, C.E. | | Deposit date: | 1994-07-18 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism.
Structure, 2, 1994
|
|
1PGN
 
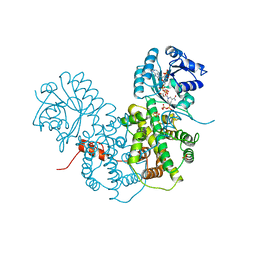 | | CRYSTALLOGRAPHIC STUDY OF COENZYME, COENZYME ANALOGUE AND SUBSTRATE BINDING IN 6-PHOSPHOGLUCONATE DEHYDROGENASE: IMPLICATIONS FOR NADP SPECIFICITY AND THE ENZYME MECHANISM | | Descriptor: | 6-PHOSPHOGLUCONATE DEHYDROGENASE, NICOTINAMIDE 8-BROMO-ADENINE DINUCLEOTIDE PHOSPHATE, PYROPHOSPHATE 2-, ... | | Authors: | Adams, M.J, Phillips, C, Gover, S. | | Deposit date: | 1994-07-18 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism.
Structure, 2, 1994
|
|
1PGQ
 
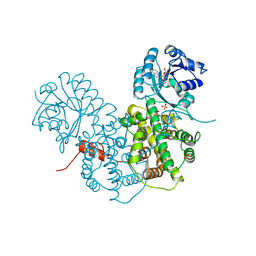 | | CRYSTALLOGRAPHIC STUDY OF COENZYME, COENZYME ANALOGUE AND SUBSTRATE BINDING IN 6-PHOSPHOGLUCONATE DEHYDROGENASE: IMPLICATIONS FOR NADP SPECIFICITY AND THE ENZYME MECHANISM | | Descriptor: | 6-PHOSPHOGLUCONATE DEHYDROGENASE, ADENOSINE-2'-MONOPHOSPHATE, SULFATE ION | | Authors: | Adams, M.J, Phillips, C, Gover, S. | | Deposit date: | 1994-07-18 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism.
Structure, 2, 1994
|
|
4QKX
 
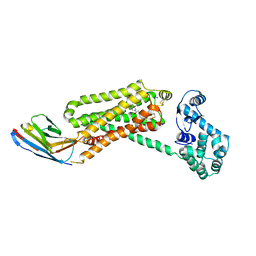 | | Structure of beta2 adrenoceptor bound to a covalent agonist and an engineered nanobody | | Descriptor: | 4-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(2-sulfanylethoxy)phenyl]ethyl}amino)ethyl]benzene-1,2-diol, Beta-2 adrenergic receptor, R9 protein, ... | | Authors: | Weichert, D, Kruse, A.C, Manglik, A, Hiller, C, Zhang, C, Huebner, H, Kobilka, B.K, Gmeiner, P. | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-23 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Covalent agonists for studying G protein-coupled receptor activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3P1M
 
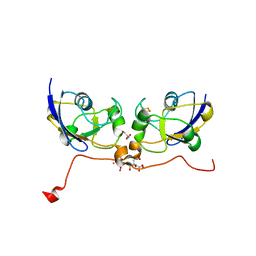 | | Crystal structure of human ferredoxin-1 (FDX1) in complex with iron-sulfur cluster | | Descriptor: | Adrenodoxin, mitochondrial, CITRATE ANION, ... | | Authors: | Chaikuad, A, Johansson, C, Krojer, T, Yue, W.W, Phillips, C, Bray, J.E, Pike, A.C.W, Muniz, J.R.C, Vollmar, M, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of human ferredoxin-1 (FDX1) in complex with iron-sulfur cluster
To be Published
|
|
3PRY
 
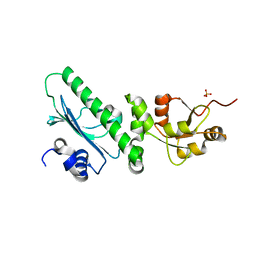 | | Crystal structure of the middle domain of human HSP90-beta refined at 2.3 A resolution | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Heat shock protein HSP 90-beta, ... | | Authors: | Chaikuad, A, Pilka, E, Sharpe, T.D, Cooper, C.D.O, Phillips, C, Berridge, G, Ayinampudi, V, Fedorov, O, Keates, T, Thangaratnarajah, C, Zimmermann, T, Vollmar, M, Yue, W.W, Che, K.H, Krojer, T, Muniz, J.R.C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-30 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the middle domain of human HSP90-beta refined at 2.3 A resolution
To be Published
|
|
6H46
 
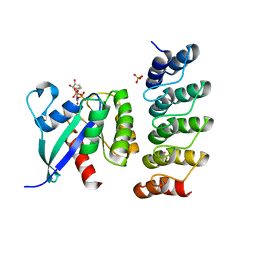 | | Human KRAS in complex with darpin K13 | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Debreczeni, J.E, Bery, N, Legg, S, Breed, J, Embrey, K, Stubbs, C, Kolasinska-Zwierz, P, Barrett, N, Marwood, R, Watson, J, Tart, J, Overman, R, Miller, A, Phillips, C, Minter, R, Rabbitts, T.H. | | Deposit date: | 2018-07-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | KRAS-specific inhibition using a DARPin binding to a site in the allosteric lobe.
Nat Commun, 10, 2019
|
|
4ZH7
 
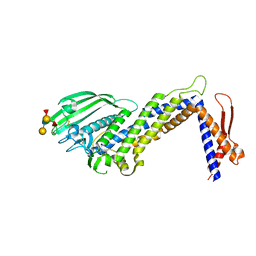 | | Structural basis of Lewisb antigen binding by the Helicobacter pylori adhesin BabA | | Descriptor: | Outer membrane protein-adhesin, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose | | Authors: | Howard, T, Hage, N, Phillips, C, Brassington, C.A, Debreczeni, J, Overman, R, Gellert, P, Stolnik, S, Winkler, G.S, Falcone, F.H. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis of Lewis(b) antigen binding by the Helicobacter pylori adhesin BabA.
Sci Adv, 1, 2015
|
|
4ZH0
 
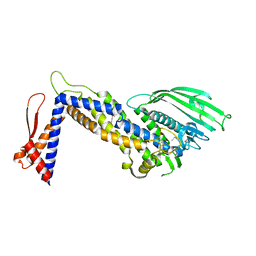 | | Structure of Helicobacter pylori adhesin BabA determined by SeMet SAD | | Descriptor: | Outer membrane protein-adhesin | | Authors: | Howard, T.D, Hage, N, Phillips, C, Brassington, C.A, Debreczeni, J, Overman, R, Gellert, P, Stolnik, S, Winkler, G.S, Falcone, F.H. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis of Lewis(b) antigen binding by the Helicobacter pylori adhesin BabA.
Sci Adv, 1, 2015
|
|
