8EHJ
 
 | |
8PYI
 
 | | Human IGF1R with inhibitor 6 | | Descriptor: | 3-[8-azanyl-1-(4-ethoxy-8-fluoranyl-2-phenyl-quinolin-7-yl)imidazo[1,5-a]pyrazin-3-yl]-1-methyl-cyclobutan-1-ol, Insulin-like growth factor 1 receptor beta chain | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Human IGF1R with inhibitor 6
To Be Published
|
|
8PYJ
 
 | | Human IGF1R with inhibitor 8 | | Descriptor: | 5,5-dimethyl-1-(quinolin-4-ylmethyl)-3-[4-(trifluoromethylsulfonyl)phenyl]imidazolidine-2,4-dione, CADMIUM ION, Insulin-like growth factor 1 receptor beta chain, ... | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Human IGF1R with inhibitor 8
To Be Published
|
|
8PYK
 
 | | Human IGF1R with inhibitor 47 | | Descriptor: | 5,5-dimethyl-1-(1H-pyrrolo[2,3-b]pyridin-3-ylmethyl)-3-[4-(trifluoromethylsulfanyl)phenyl]imidazolidine-2,4-dione, Insulin-like growth factor 1 receptor beta chain, NICKEL (II) ION | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Human IGF1R with inhibitor 47
To Be Published
|
|
8PYL
 
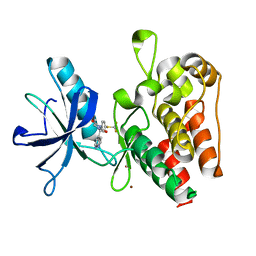 | | Human IGF1R with inhibitor 53 | | Descriptor: | (5S)-5-methyl-1-(1H-pyrrolo[2,3-b]pyridin-4-ylmethyl)-3-[4-(trifluoromethylsulfanyl)phenyl]imidazolidine-2,4-dione, CADMIUM ION, Insulin-like growth factor 1 receptor beta chain, ... | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Human IGF1R with inhibitor 53
To Be Published
|
|
8PYN
 
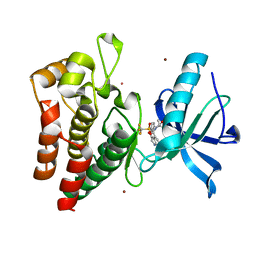 | | Human IGF1R with inhibitor 56 | | Descriptor: | 5,5-dimethyl-1-(1H-pyrrolo[2,3-b]pyridin-4-ylmethyl)-3-[4-(trifluoromethylsulfonyl)phenyl]imidazolidine-2,4-dione, CADMIUM ION, Insulin-like growth factor 1 receptor beta chain, ... | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Human IGF1R with inhibitor 56
To Be Published
|
|
8PYM
 
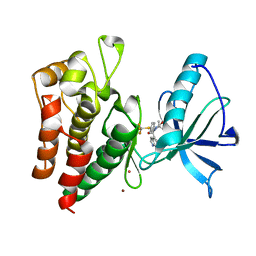 | | Human IGF1R with inhibitor 54 | | Descriptor: | 5,5-dimethyl-1-(1H-pyrrolo[2,3-b]pyridin-4-ylmethyl)-3-[4-(trifluoromethylsulfanyl)phenyl]imidazolidine-2,4-dione, CADMIUM ION, Insulin-like growth factor 1 receptor beta chain, ... | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Human IGF1R with inhibitor 54
To Be Published
|
|
8FTL
 
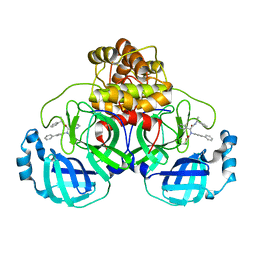 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease (Mpro) in complex with inhibitor Jun89-3-C1 | | Descriptor: | 3C-like proteinase nsp5, N-([1,1'-biphenyl]-4-yl)-2-chloro-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]acetamide | | Authors: | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of the SARS-CoV-2 (COVID-19) main protease (Mpro) in complex with inhibitor Jun89-3-C1
To Be Published
|
|
7YOJ
 
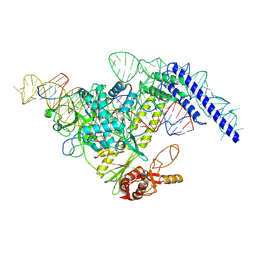 | | Structure of CasPi with guide RNA and target DNA | | Descriptor: | CasPi, DNA (30-MER), DNA (5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*CP*CP*AP*G)-3'), ... | | Authors: | Li, C.P, Wang, J, Liu, J.J. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | The compact Cas pi (Cas12l) 'bracelet' provides a unique structural platform for DNA manipulation.
Cell Res., 33, 2023
|
|
6LOX
 
 | | Crystal Structure of human glutaminase with macrocyclic inhibitor | | Descriptor: | (E)-15,22-Dioxa-4,11-diaza-5(2,5)-thiadiazola-10(3,6)-pyridazina-1,14(1,3)-dibenzenacyclodocosaphan-18-ene-3,12-dione, Glutaminase kidney isoform, mitochondrial | | Authors: | Bian, J, Li, Z, Xu, X, Wang, J, Li, L. | | Deposit date: | 2020-01-07 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Enabled Discovery of Novel Macrocyclic Inhibitors Targeting Glutaminase 1 Allosteric Binding Site.
J.Med.Chem., 64, 2021
|
|
5X40
 
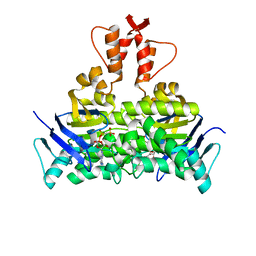 | | Structure of a CbiO dimer bound with AMPPCP | | Descriptor: | Cobalt ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Bao, Z, Qi, X, Wang, J, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
4N77
 
 | |
7DAC
 
 | | Human RIPK3 amyloid fibril revealed by solid-state NMR | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Wu, X.L, Zhang, J, Dong, X.Q, Liu, J, Li, B, Hu, H, Wang, J, Wang, H.Y, Lu, J.X. | | Deposit date: | 2020-10-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7D8T
 
 | | MITF bHLHLZ complex with M-box DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*AP*CP*AP*TP*GP*TP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*AP*CP*AP*CP*AP*TP*GP*TP*TP*AP*CP*AP*G)-3'), Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8R
 
 | | MITF HLHLZ structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8S
 
 | | MITF bHLHLZ apo structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit, SULFATE ION | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
5XP8
 
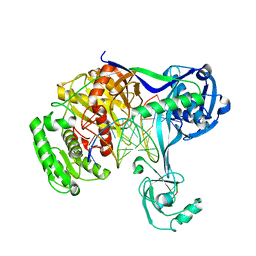 | | Crystal structure of T. thermophilus Argonaute protein complexed with a bulge 4A5 on the guide strand | | Descriptor: | DNA (5'-D(*AP*GP*T)-3'), DNA (5'-D(*CP*AP*AP*CP*C*AP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3'), DNA (5'-D(P*TP*GP*AP*GP*AP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*T)-3'), ... | | Authors: | Sheng, G, Gogakos, T, Wang, J, Zhao, H, Serganov, A, Juranek, S, Tuschl, T, Patel, J.D, Wang, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes.
Nucleic Acids Res., 45, 2017
|
|
7DDP
 
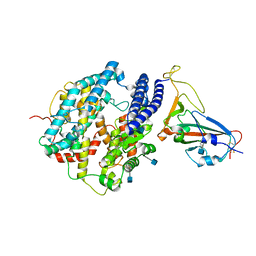 | | Cryo-EM structure of human ACE2 and GX/P2V/2017 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Niu, S, Wang, J, Wang, H.W, Qi, J.X, Wang, Q.H, Gao, G.F. | | Deposit date: | 2020-10-29 | | Release date: | 2021-05-19 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin.
Embo J., 40, 2021
|
|
7DDO
 
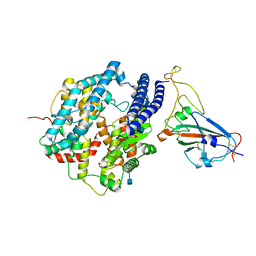 | | Cryo-EM structure of human ACE2 and GD/1/2019 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Niu, S, Wang, J, Wang, H.W, Qi, J.X, Wang, Q.H, Gao, G.F. | | Deposit date: | 2020-10-29 | | Release date: | 2021-05-19 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin.
Embo J., 40, 2021
|
|
7D0F
 
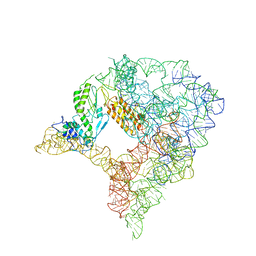 | | cryo-EM structure of a pre-catalytic group II intron RNP | | Descriptor: | Group II intron-encoded protein LtrA, RNA (738-MER) | | Authors: | Liu, N, Dong, X.L, Hu, C.X, Zeng, J.W, Wang, J.W, Wang, J, Wang, H.W, Belfort, M. | | Deposit date: | 2020-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Exon and protein positioning in a pre-catalytic group II intron RNP primed for splicing.
Nucleic Acids Res., 48, 2020
|
|
7D0G
 
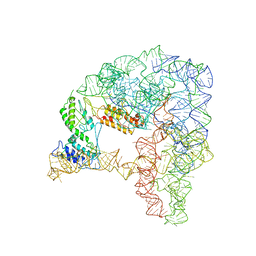 | | Cryo-EM structure of a pre-catalytic group II intron | | Descriptor: | Group II intron-encoded protein LtrA, RNA (714-MER) | | Authors: | Liu, N, Dong, X.L, Hu, C.X, Zeng, J.W, Wang, J.W, Wang, J, Wang, H.W, Belfort, M. | | Deposit date: | 2020-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Exon and protein positioning in a pre-catalytic group II intron RNP primed for splicing.
Nucleic Acids Res., 48, 2020
|
|
7D1A
 
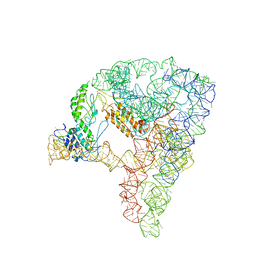 | | cryo-EM structure of a group II intron RNP complexed with its reverse transcriptase | | Descriptor: | Group II intron-encoded protein LtrA, RNA (5'-R(P*CP*AP*CP*AP*UP*CP*CP*AP*UP*AP*AP*C)-3'), RNA (692-MER) | | Authors: | Liu, N, Dong, X.L, Hu, C.X, Zeng, J.W, Wang, J.W, Wang, J, Wang, H.W, Belfort, M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Exon and protein positioning in a pre-catalytic group II intron RNP primed for splicing.
Nucleic Acids Res., 48, 2020
|
|
7YVR
 
 | | Crystal Structure of L-Threonine Aldolase from Neptunomonas marina | | Descriptor: | GLYCEROL, L-threonine aldolase | | Authors: | He, Y.Z, Wang, J, Yan, W.P, Zhang, Y, Feng, Y. | | Deposit date: | 2022-08-19 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Engineering of the L-Threonine Aldolase from Neptunomonas marine for the Efficient Synthesis of beta-Hydroxy-alpha-amino Acids via C-C Formation
Acs Catalysis, 2023
|
|
4MM2
 
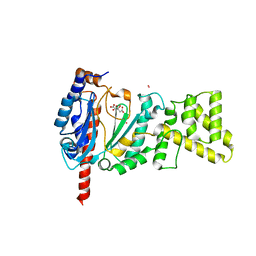 | | Crystal structure of yeast primase catalytic subunit | | Descriptor: | CADMIUM ION, CITRIC ACID, DNA primase small subunit | | Authors: | Park, K.R, An, J.Y, Lee, Y, Youn, H.S, Lee, J.G, Kang, J.Y, Kim, T.G, Lim, J.J, Eom, S.H, Wang, J. | | Deposit date: | 2013-09-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of yeast primase catalytic subunit
To be Published
|
|
7D73
 
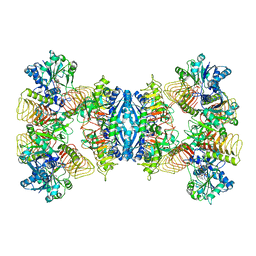 | | Cryo-EM structure of GMPPA/GMPPB complex bound to GTP (State I) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, GUANOSINE-5'-TRIPHOSPHATE, Mannose-1-phosphate guanyltransferase alpha, ... | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, Cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
