7EU5
 
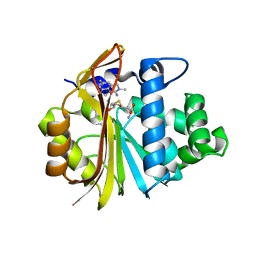 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with tricyclic small molecule inhibitor JBSNF-000107 | | Descriptor: | 6-fluoranyl-10-methyl-1,10-diazatricyclo[6.3.1.0^{4,12}]dodeca-4,6,8(12)-trien-11-imine, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Swaminathan, S, Gosu, R, Birudukota, S, Kandan, S, Vaithilingam, K. | | Deposit date: | 2021-05-16 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.731 Å) | | Cite: | Novel tricyclic small molecule inhibitors of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 12, 2022
|
|
7ET7
 
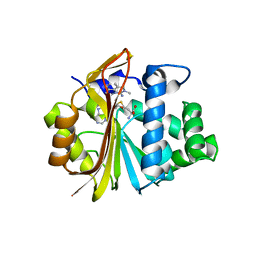 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with tricyclic small molecule inhibitor JBSNF-000028 | | Descriptor: | 10-methyl-1,10-diazatricyclo[6.3.1.0^{4,12}]dodeca-4,6,8(12)-trien-11-imine, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Swaminathan, S, Gosu, R, Birudukota, S, Kandan, S, Vaithilingam, K. | | Deposit date: | 2021-05-12 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Novel tricyclic small molecule inhibitors of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 12, 2022
|
|
3PJI
 
 | |
2ETF
 
 | |
2EUI
 
 | |
1SIX
 
 | | Mycobacterium tuberculosis dUTPase complexed with magnesium and alpha,beta-imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.-S, Kim, M, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
1SNF
 
 | | MYCOBACTERIUM TUBERCULOSIS DUTPASE COMPLEXED WITH MAGNESIUM AND DEOXYURIDINE 5'-MONOPHOSPHATE | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-10 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
1SM8
 
 | | M. tuberculosis dUTPase complexed with chromium and dUTP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHROMIUM ION, DEOXYURIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-08 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
1SJN
 
 | | Mycobacterium tuberculosis dUTPase complexed with magnesium and alpha,beta-imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-04 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
1SMC
 
 | | Mycobacterium tuberculosis dUTPase complexed with dUTP in the absence of metal ion. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE-5'-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
1SLH
 
 | | Mycobacterium tuberculosis dUTPase complexed with magnesium and dUDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-05 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
2OG2
 
 | | Crystal structure of chloroplast FtsY from Arabidopsis thaliana | | Descriptor: | MAGNESIUM ION, MALONATE ION, Putative signal recognition particle receptor | | Authors: | Chartron, J, Chandrasekar, S, Ampornpan, P.J, Shan, S. | | Deposit date: | 2007-01-04 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Chloroplast Signal Recognition Particle (SRP) Receptor: Domain Arrangement Modulates SRP-Receptor Interaction.
J.Mol.Biol., 375, 2007
|
|
3DOX
 
 | | X-ray structure of HIV-1 protease in situ product complex | | Descriptor: | A PEPTIDE SUBSTRATE-PIV, A PEPTIDE SUBSTRATE-SQNY, HIV-1 PROTEASE | | Authors: | Hosur, M.V, Ferrer, J.-L, Das, A, Prashar, V, Bihani, S. | | Deposit date: | 2008-07-07 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of HIV-1 protease in situ product complex
Proteins, 74, 2009
|
|
1VFC
 
 | | Solution Structure Of The DNA Complex Of Human Trf2 | | Descriptor: | Short C-rich starnd, Short G-rich strand, Telomeric repeat binding factor 2 | | Authors: | Nishimura, Y, Hanaoka, S. | | Deposit date: | 2004-04-12 | | Release date: | 2005-05-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities
Protein Sci., 14, 2005
|
|
1VF9
 
 | | Solution Structure Of Human Trf2 | | Descriptor: | Telomeric repeat binding factor 2 | | Authors: | Nishimura, Y, Hanaoka, S. | | Deposit date: | 2004-04-12 | | Release date: | 2005-05-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities
Protein Sci., 14, 2005
|
|
2J5A
 
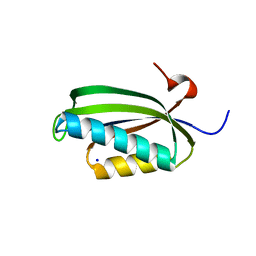 | | Folding of S6 structures with divergent amino-acid composition: pathway flexibility within partly overlapping foldons | | Descriptor: | 30S RIBOSOMAL PROTEIN S6, SODIUM ION | | Authors: | Hansson, S, Olofsson, L, Hedberg, L, Oliveberg, M, Logan, D.T. | | Deposit date: | 2006-09-13 | | Release date: | 2006-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Folding of S6 Structures with Divergent Amino Acid Composition: Pathway Flexibility within Partly Overlapping Foldons.
J.Mol.Biol., 365, 2007
|
|
3Q4H
 
 | | Crystal structure of the Mycobacterium smegmatis EsxGH complex (MSMEG_0620-MSMEG_0621) | | Descriptor: | Low molecular weight protein antigen 7, Pe family protein | | Authors: | Chan, S, Harris, L, Kuo, E, Ahn, C, Zhou, T.T, Nguyen, L, Shin, A, Sawaya, M.R, Cascio, D, Arbing, M.A, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-12-23 | | Release date: | 2011-01-26 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
4PG3
 
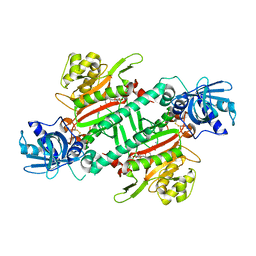 | | Crystal structure of KRS complexed with inhibitor | | Descriptor: | LYSINE, Lysine--tRNA ligase, cladosporin | | Authors: | Sharma, A, Yogavel, M, Khan, S, Sharma, A, Belrhali, H. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Structural basis of malaria parasite lysyl-tRNA synthetase inhibition by cladosporin.
J. Struct. Funct. Genomics, 15, 2014
|
|
5OWF
 
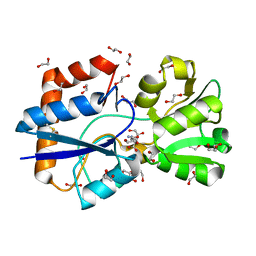 | | Structure of a LAO-binding protein mutant with glutamine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLUTAMINE, ... | | Authors: | Shanmugaratnam, S, Banda-Vazquez, J, Sosa-Peinado, A, Hocker, B. | | Deposit date: | 2017-08-31 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Redesign of LAOBP to bind novel l-amino acid ligands.
Protein Sci., 27, 2018
|
|
7WHU
 
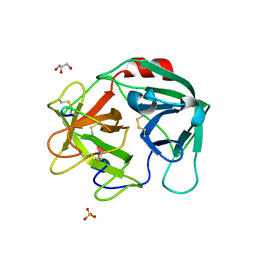 | | Human Neutrophil Elastase in-complex with Ecotin Peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ecotin Peptide, ... | | Authors: | Shankar, S, Jayaraman, S. | | Deposit date: | 2021-12-31 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Sequence preference and scaffolding requirement for the inhibition of human neutrophil elastase by ecotin peptide
Protein Sci., 31, 2022
|
|
7FCD
 
 | | Structure of the SARS-CoV-2 A372T spike glycoprotein (open) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Zhang, S. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-26 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Loss of Spike N370 glycosylation as an important evolutionary event for the enhanced infectivity of SARS-CoV-2.
Cell Res., 32, 2022
|
|
7FCE
 
 | | Structure of the SARS-CoV-2 A372T spike glycoprotein (closed) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Zhang, S. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-26 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Loss of Spike N370 glycosylation as an important evolutionary event for the enhanced infectivity of SARS-CoV-2.
Cell Res., 32, 2022
|
|
2RHB
 
 | | Crystal structure of Nsp15-H234A mutant- Hexamer in asymmetric unit | | Descriptor: | Uridylate-specific endoribonuclease | | Authors: | Palaninathan, S, Bhardwaj, K, Alcantara, J.M.O, Guarino, L, Yi, L.L, Kao, C.C, Sacchettini, J. | | Deposit date: | 2007-10-08 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15.
J.Biol.Chem., 283, 2008
|
|
7VSO
 
 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles in Complex with HAD16 Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | 2-[[(2R)-2-(3-bromanyl-5-iodanyl-phenyl)carbonyloxy-3-tetradecanoyloxy-propoxy]-oxidanyl-phosphoryl]oxyethyl-trimethyl-azanium, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Bacteriorhodopsin, ... | | Authors: | Mizohata, E, Nakane, T, Hanashima, S. | | Deposit date: | 2021-10-27 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Heavy Atom Detergent/Lipid Combined X-ray Crystallography for Elucidating the Structure-Function Relationships of Membrane Proteins.
Membranes (Basel), 11, 2021
|
|
7WB0
 
 | |
