4U9P
 
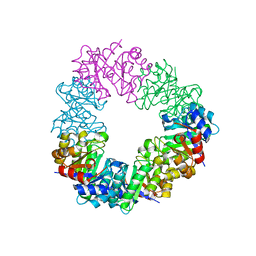 | | Structure of the methanofuran/methanopterin biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii | | Descriptor: | GLYCEROL, UPF0264 protein MJ1099 | | Authors: | Bobik, T.A, Morales, E, Shin, A, Cascio, D, Sawaya, M.R, Arbing, M, Rasche, M.E. | | Deposit date: | 2014-08-06 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the methanofuran/methanopterin-biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3U2H
 
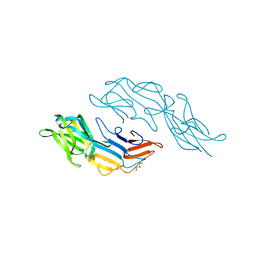 | | Crystal structure of the C-terminal DUF1608 domain of the Methanosarcina acetivorans S-layer (MA0829) protein | | Descriptor: | GLYCEROL, S-layer protein MA0829 | | Authors: | Chan, S, Phan, T, Ahn, C.J, Shin, A, Rohlin, L, Gunsalus, R.P, Arbing, M.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3U2G
 
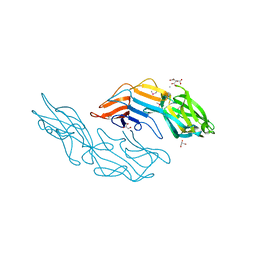 | | Crystal structure of the C-terminal DUF1608 domain of the Methanosarcina acetivorans S-layer (MA0829) protein | | Descriptor: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Chan, S, Phan, T, Ahn, C.J, Shin, A, Rohlin, L, Gunsalus, R.P, Arbing, M.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3MML
 
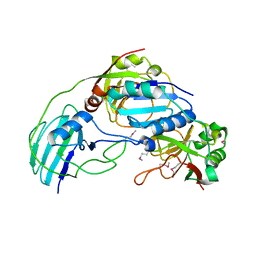 | | Allophanate Hydrolase Complex from Mycobacterium smegmatis, Msmeg0435-Msmeg0436 | | Descriptor: | Allophanate hydrolase subunit 1, Allophanate hydrolase subunit 2, CHLORIDE ION | | Authors: | Kaufmann, M, Chernishof, I, Shin, A, Germano, D, Sawaya, M.R, Waldo, G.S, Arbing, M.A, Perry, J, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Allphanate Hydrolase Complex from M. smegmatis, Msmeg0435-Msmeg0436
To be Published
|
|
3Q4H
 
 | | Crystal structure of the Mycobacterium smegmatis EsxGH complex (MSMEG_0620-MSMEG_0621) | | Descriptor: | Low molecular weight protein antigen 7, Pe family protein | | Authors: | Chan, S, Harris, L, Kuo, E, Ahn, C, Zhou, T.T, Nguyen, L, Shin, A, Sawaya, M.R, Cascio, D, Arbing, M.A, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-12-23 | | Release date: | 2011-01-26 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
5DBN
 
 | | Crystal structure of AtoDA complex | | Descriptor: | Acetate CoA-transferase subunit alpha, Acetate CoA-transferase subunit beta, CHLORIDE ION, ... | | Authors: | Arbing, M.A, Koo, C.W, Shin, A, Medrano-Soto, A, Eisenberg, D. | | Deposit date: | 2015-08-21 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Crystal structure of AtoDA complex
To Be Published
|
|
5DB5
 
 | | Crystal structure of PLP-bound E. coli SufS (cysteine persulfide intermediate) in space group P21 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, CYSTEINE, ... | | Authors: | Arbing, M.A, Shin, A, Koo, C.W, Medrano-Soto, A, Eisenberg, D. | | Deposit date: | 2015-08-20 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of PLP-bound E. coli SufS (cysteine persulfide intermediate) in space group P21
To Be Published
|
|
5DUD
 
 | | Crystal structure of E. coli YbgJK | | Descriptor: | YbgJ, YbgK | | Authors: | Arbing, M.A, Kaufmann, M, Shin, A, Medrano-Soto, A, Cascio, D, Eisenberg, D. | | Deposit date: | 2015-09-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of E. coli YbgJK
To Be Published
|
|
5HQL
 
 | | Structure function studies of R. palustris RubisCO (A47V-M331A mutant; CABP-bound; no expression tag) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Shin, A, Cascio, D, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure function studies of R. palustris RubisCO
To Be Published
|
|
5HAO
 
 | | Structure function studies of R. palustris RubisCO (M331A mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Shin, A, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2015-12-30 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
5HJY
 
 | | Structure function studies of R. palustris RubisCO (I165T mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Arbing, M.A, Shin, A, Cascio, D, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2016-01-13 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
5HJX
 
 | | Structure function studies of R. palustris RubisCO (A47V mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Shin, A, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2016-01-13 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
5HK4
 
 | | Structure function studies of R. palustris RubisCO (A47V-M331A mutant) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Shin, A, Cascio, D, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2016-01-13 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
5HQM
 
 | | Structure function studies of R. palustris RubisCO (R. palustris/R. rubrum chimera) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Arbing, M.A, Shin, A, Cascio, D, Satagopan, S, Tabita, F.R. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
2NCY
 
 | | Solution structure of pseudin-2 analog (Ps-P) | | Descriptor: | Pseudin-2 | | Authors: | Jeon, D, Kim, J, Shin, A, Kim, Y. | | Deposit date: | 2016-04-18 | | Release date: | 2017-04-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Optimum Balance between the Cationicity and Structural Component for Bacterial Cell Selectivity and Anti-inflammatory activities of Pseudin-2 and its Analogs
To be Published
|
|
2NCX
 
 | |
