7N7Z
 
 | |
2H28
 
 | | Crystal structure of YeeU from E. coli. Northeast Structural Genomics target ER304 | | Descriptor: | CHLORIDE ION, GLYCEROL, Hypothetical protein yeeU, ... | | Authors: | Arbing, M, Su, M, Benach, J, Karpowich, N.K, Jiang, M, Xiao, R, Cunningham, K, Ma, L.-C, Chen, C.X, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-18 | | Release date: | 2006-07-18 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
4I0X
 
 | | Crystal structure of the Mycobacterum abscessus EsxEF (Mab_3112-Mab_3113) complex | | Descriptor: | BETA-MERCAPTOETHANOL, ESAT-6-like protein MAB_3112, ESAT-6-like protein MAB_3113, ... | | Authors: | Arbing, M.A, Chan, S, Lu, J, Kuo, E, Harris, L, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-11-19 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
4GZR
 
 | | Crystal structure of the Mycobacterium tuberculosis H37Rv EsxOP (Rv2346c-Rv2347c) complex in space group C2221 | | Descriptor: | ESAT-6-like protein 6, ESAT-6-like protein 7, SULFATE ION | | Authors: | Arbing, M.A, Chan, S, He, Q, Harris, L, Zhou, T.T, Kuo, E, Ahn, C.J, Eisenberg, D, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2012-09-06 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
8TFS
 
 | |
2ICP
 
 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 4.0. Northeast Structural Genomics Consortium TARGET ER390. | | Descriptor: | MAGNESIUM ION, antitoxin higa | | Authors: | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the bacterial antitoxin HigA from Escherichia coli.
To be Published
|
|
2ICT
 
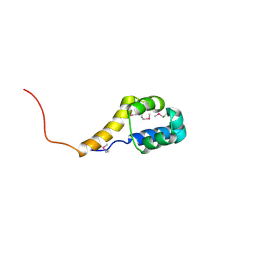 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 8.5. Northeast Structural Genomics TARGET ER390. | | Descriptor: | antitoxin higa | | Authors: | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
3OGI
 
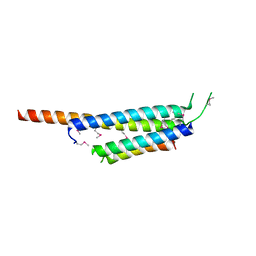 | | Crystal structure of the Mycobacterium tuberculosis H37Rv EsxOP complex (Rv2346c-Rv2347c) | | Descriptor: | Putative ESAT-6-like protein 6, Putative ESAT-6-like protein 7 | | Authors: | Arbing, M.A, Chan, S, Zhou, T.T, Ahn, C, Harris, L, Kuo, E, Sawaya, M.R, Cascio, D, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-08-16 | | Release date: | 2010-08-25 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
3KH2
 
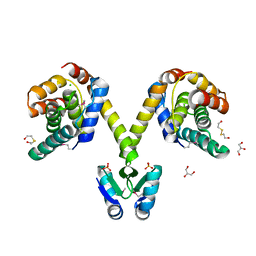 | | Crystal structure of the P1 bacteriophage Doc toxin (F68S) in complex with the Phd antitoxin (L17M/V39A). Northeast Structural Genomics targets ER385-ER386 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Death on curing protein, ... | | Authors: | Arbing, M.A, Kuzin, A.P, Su, M, Abashidze, M, Verdon, G, Liu, M, Xiao, R, Acton, T, Inouye, M, Montelione, G.T, Woychik, N.A, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-29 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
5KOZ
 
 | | Structure function studies of R. palustris RubisCO (K192C mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, CARBONATE ION, MAGNESIUM ION, ... | | Authors: | Arbing, M.A, North, J.A, Satagopan, S, Tabita, F.R. | | Deposit date: | 2016-07-01 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
4PM4
 
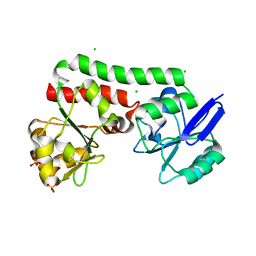 | | Structure of a putative periplasmic iron siderophore binding protein (Rv0265c) from Mycobacterium tuberculosis H37Rv | | Descriptor: | CHLORIDE ION, Iron complex transporter substrate-binding protein, SULFATE ION | | Authors: | Arbing, M.A, Chan, S, Tran, N, Kuo, E, Lu, J, Harris, L.R, Zhou, T.T, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-05-20 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a putative periplasmic iron siderophore binding protein (Rv0265c) from Mycobacterium tuberculosis H37Rv
To Be Published
|
|
4ZJM
 
 | | Crystal Structure of Mycobacterium tuberculosis LpqH (Rv3763) | | Descriptor: | CHLORIDE ION, GLYCEROL, Lipoprotein LpqH, ... | | Authors: | Arbing, M.A, Chan, S, Kuo, E, Harris, L.R, Zhou, T.T, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Crystal Structure of Mycobacterium tuberculosis LpqH (Rv3763)
To Be Published
|
|
5HQM
 
 | | Structure function studies of R. palustris RubisCO (R. palustris/R. rubrum chimera) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Arbing, M.A, Shin, A, Cascio, D, Satagopan, S, Tabita, F.R. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
4RZT
 
 | | Lac repressor engineered to bind sucralose, sucralose-bound tetramer | | Descriptor: | 4-chloro-4-deoxy-alpha-D-galactopyranose-(1-2)-1,6-dichloro-1,6-dideoxy-beta-D-fructofuranose, Lac repressor | | Authors: | Arbing, M.A, Cascio, D, Sawaya, M.R, Kosuri, S, Church, G.M. | | Deposit date: | 2014-12-24 | | Release date: | 2015-12-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Engineering an allosteric transcription factor to respond to new ligands.
Nat.Methods, 13, 2016
|
|
5HAT
 
 | | Structure function studies of R. palustris RubisCO (S59F/M331A mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Leong, J.G, Cascio, D, Varaljay, V.A, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2015-12-30 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
5HK4
 
 | | Structure function studies of R. palustris RubisCO (A47V-M331A mutant) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Shin, A, Cascio, D, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2016-01-13 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
5HAN
 
 | | Structure function studies of R. palustris RubisCO (S59F mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Leong, J.G, Varaljay, V.A, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2015-12-30 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure function studies of R. palustris RubisCO
To Be Published
|
|
5HAO
 
 | | Structure function studies of R. palustris RubisCO (M331A mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Shin, A, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2015-12-30 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
5HJY
 
 | | Structure function studies of R. palustris RubisCO (I165T mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Arbing, M.A, Shin, A, Cascio, D, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2016-01-13 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
5HJX
 
 | | Structure function studies of R. palustris RubisCO (A47V mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Shin, A, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2016-01-13 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
4RZS
 
 | | Lac repressor engineered to bind sucralose, unliganded tetramer | | Descriptor: | GLYCEROL, Lac repressor | | Authors: | Arbing, M.A, Cascio, D, Kosuri, S, Church, G.M. | | Deposit date: | 2014-12-24 | | Release date: | 2015-12-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Engineering an allosteric transcription factor to respond to new ligands.
Nat.Methods, 13, 2016
|
|
5HQL
 
 | | Structure function studies of R. palustris RubisCO (A47V-M331A mutant; CABP-bound; no expression tag) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Shin, A, Cascio, D, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure function studies of R. palustris RubisCO
To Be Published
|
|
5C2C
 
 | | GWS1B RubisCO: Form II RubisCO derived from uncultivated Gallionellacea species (unliganded form) | | Descriptor: | CHLORIDE ION, Form II RubisCO, MAGNESIUM ION | | Authors: | Arbing, M.A, Varaljay, V.A, Satagopan, S, Tabita, F.R. | | Deposit date: | 2015-06-15 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Functional metagenomic selection of ribulose 1, 5-bisphosphate carboxylase/oxygenase from uncultivated bacteria.
Environ.Microbiol., 18, 2016
|
|
5C2G
 
 | | GWS1B RubisCO: Form II RubisCO derived from uncultivated Gallionellacea species (CABP-bound). | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, Form II RubisCO, MAGNESIUM ION | | Authors: | Arbing, M.A, Varaljay, V.A, Satagopan, S, Tabita, F.R. | | Deposit date: | 2015-06-15 | | Release date: | 2015-12-16 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Functional metagenomic selection of ribulose 1, 5-bisphosphate carboxylase/oxygenase from uncultivated bacteria.
Environ.Microbiol., 18, 2016
|
|
5DUD
 
 | | Crystal structure of E. coli YbgJK | | Descriptor: | YbgJ, YbgK | | Authors: | Arbing, M.A, Kaufmann, M, Shin, A, Medrano-Soto, A, Cascio, D, Eisenberg, D. | | Deposit date: | 2015-09-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of E. coli YbgJK
To Be Published
|
|
