7Y4T
 
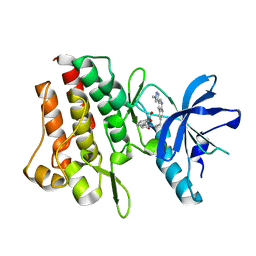 | | Crystal structure of cMET kinase domain bound by compound 9I | | Descriptor: | 2-[2-[3-(1-methylpyrazol-4-yl)quinolin-6-yl]ethyl]-6-(3-nitrophenyl)pyridazin-3-one, Hepatocyte growth factor receptor | | Authors: | Qu, L.Z, Chen, Y.H. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of D6808, a Highly Selective and Potent Macrocyclic c-Met Inhibitor for Gastric Cancer Harboring MET Gene Alteration Treatment.
J.Med.Chem., 65, 2022
|
|
7Y4U
 
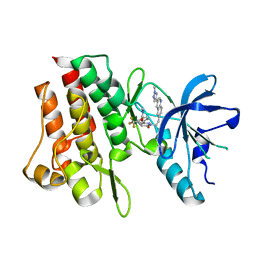 | | Crystal structure of cMET kinase domain bound by compound 9Y | | Descriptor: | Hepatocyte growth factor receptor, ~{N}-methyl-4-[1-[2-[3-(1-methylpyrazol-4-yl)quinolin-6-yl]ethyl]-6-oxidanylidene-pyridazin-3-yl]-2-(trifluoromethyl)benzamide | | Authors: | Qu, L.Z, Chen, Y.H. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of D6808, a Highly Selective and Potent Macrocyclic c-Met Inhibitor for Gastric Cancer Harboring MET Gene Alteration Treatment.
J.Med.Chem., 65, 2022
|
|
7YPX
 
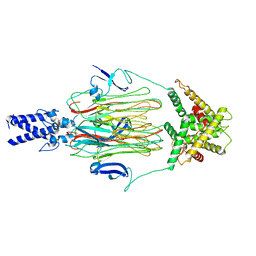 | | Cyanophage Pam3 fiber | | Descriptor: | Pam3 tail fiber proreins, tail fiber chaperone | | Authors: | Wei, Z.L, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-08-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural Insights into the Chaperone-Assisted Assembly of a Simplified Tail Fiber of the Myocyanophage Pam3.
Viruses, 14, 2022
|
|
7YQK
 
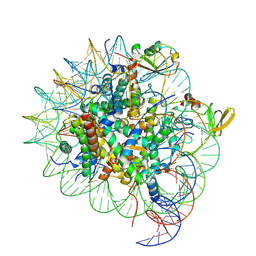 | | cryo-EM structure of gammaH2AXK15ub-H4K20me2 nucleosome bound to 53BP1 | | Descriptor: | DNA (145-MER), Histone H2AX, Histone H2B, ... | | Authors: | Ai, H.S, GuoChao, C, Qingyue, G, Ze-Bin, T, Zhiheng, D, Xin, L, Fan, Y, Ziyu, X, Jia-Bin, L, Changlin, T, Liu, L. | | Deposit date: | 2022-08-07 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Chemical Synthesis of Post-Translationally Modified H2AX Reveals Redundancy in Interplay between Histone Phosphorylation, Ubiquitination, and Methylation on the Binding of 53BP1 with Nucleosomes.
J.Am.Chem.Soc., 144, 2022
|
|
7YTU
 
 | |
7YTT
 
 | |
8IFP
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 1 | | Descriptor: | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFT
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 10 | | Descriptor: | (8S)-N-[(1S)-1-cyano-2-[(3S)-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2S)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFQ
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFS
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 7 | | Descriptor: | (8~{S})-7-[(2~{S})-2-(~{tert}-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFR
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3 | | Descriptor: | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-5-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]hex-3-en-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IGX
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 9 (simnotrelvir, SIM0417, SSD8432) | | Descriptor: | (8~{S})-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2~{S})-3,3-dimethyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IGY
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
7WKS
 
 | | Apo state of AtPIN3 | | Descriptor: | Auxin efflux carrier component 3 | | Authors: | Su, N. | | Deposit date: | 2022-01-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
7WKW
 
 | | NPA bound state of AtPIN3 | | Descriptor: | 2-(naphthalen-1-ylcarbamoyl)benzoic acid, Auxin efflux carrier component 3 | | Authors: | Su, N. | | Deposit date: | 2022-01-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
7WN8
 
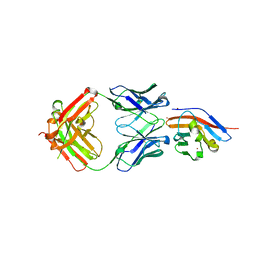 | | Crystal structure of antibody (BC31M5) binds to CD47 | | Descriptor: | BC31M5 Fab Heavy chain, BC31M5 Fab Light chain, Leukocyte surface antigen CD47, ... | | Authors: | Li, Y, Wang, W, Sui, J, Zhang, S. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A pH-dependent anti-CD47 antibody that selectively targets solid tumors and improves therapeutic efficacy and safety.
J Hematol Oncol, 16, 2023
|
|
8WPG
 
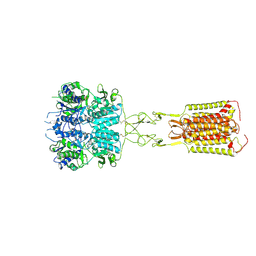 | | Human calcium-sensing receptor bound with cinacalcet in detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ling, S.L, Meng, X.Y, Tian, C.L. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into asymmetric activation of the calcium-sensing receptor-G q complex.
Cell Res., 34, 2024
|
|
8WPU
 
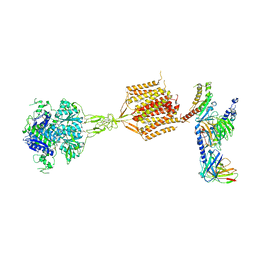 | | Human calcium-sensing receptor(CaSR) bound to cinacalcet in complex with Gq protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ling, S.L, Meng, X.Y, Tian, C.L. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into asymmetric activation of the calcium-sensing receptor-G q complex.
Cell Res., 34, 2024
|
|
7FDV
 
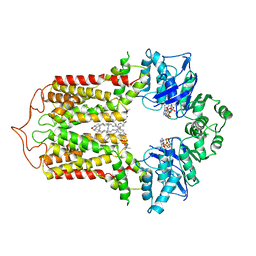 | | Cryo-EM structure of the human cholesterol transporter ABCG1 in complex with cholesterol | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family G member 1, CHOLESTEROL, ... | | Authors: | Xu, D, Li, Y.Y, Yang, F.R, Sun, C.R, Pan, J.H, Wang, L, Chen, Z.P, Fang, S.C, Yao, X.B, Hou, W.T, Zhou, C.Z, Chen, Y. | | Deposit date: | 2021-07-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure and transport mechanism of the human cholesterol transporter ABCG1.
Cell Rep, 38, 2022
|
|
1J4V
 
 | | CYANOVIRIN-N | | Descriptor: | CYANOVIRIN-N | | Authors: | Clore, G.M, Bewley, C.A. | | Deposit date: | 2001-11-21 | | Release date: | 2002-03-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Using conjoined rigid body/torsion angle simulated annealing to determine the relative orientation of covalently linked protein domains from dipolar couplings.
J.Magn.Reson., 154, 2002
|
|
7EQ1
 
 | | GPR114-Gs-scFv16 complex | | Descriptor: | Adhesion G-protein coupled receptor G5, Gs protein alpha subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ping, Y. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the tethered peptide activation of adhesion GPCRs.
Nature, 604, 2022
|
|
5XA6
 
 | |
6KTR
 
 | | Crystal structure of fibroblast growth factor 19 in complex with Fab | | Descriptor: | Fibroblast growth factor 19, G1A8-Fab-HC, G1A8-Fab-LC, ... | | Authors: | Liu, H, Zheng, S, Hou, X, Liu, X, Lv, X, Li, Y, Li, W, Sui, J. | | Deposit date: | 2019-08-28 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59775758 Å) | | Cite: | Novel Abs targeting the N-terminus of fibroblast growth factor 19 inhibit hepatocellular carcinoma growth without bile-acid-related side-effects.
Cancer Sci., 111, 2020
|
|
6JL7
 
 | |
6AFR
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with 5-((4-fluoro-1H-imidazol-1-yl)methyl)quinolin-8-ol | | Descriptor: | 5-[(4-fluoranylimidazol-1-yl)methyl]quinolin-8-ol, Bromodomain-containing protein 4 | | Authors: | Xing, J, Zhang, R.K, Zheng, M.Y, Luo, C, Jiang, X.R. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Rational design of 5-((1H-imidazol-1-yl)methyl)quinolin-8-ol derivatives as novel bromodomain-containing protein 4 inhibitors
Eur J Med Chem, 163, 2018
|
|
