3I2T
 
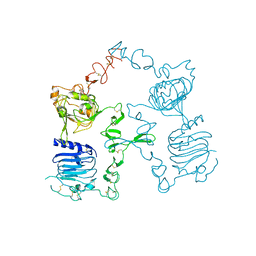 | | Crystal structure of the unliganded Drosophila Epidermal Growth Factor Receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Alvarado, D, Klein, D.E, Lemmon, M.A. | | Deposit date: | 2009-06-29 | | Release date: | 2009-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ErbB2 resembles an autoinhibited invertebrate epidermal growth factor receptor.
Nature, 461, 2009
|
|
3LTG
 
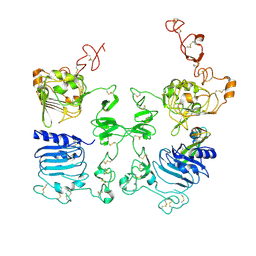 | |
3LTF
 
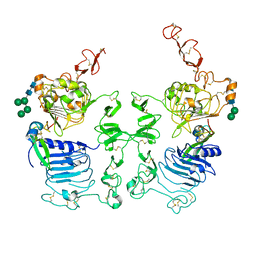 | | Crystal Structure of the Drosophila Epidermal Growth Factor Receptor ectodomain in complex with Spitz | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Alvarado, D, Klein, D.E, Lemmon, M.A. | | Deposit date: | 2010-02-15 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for negative cooperativity in growth factor binding to an EGF receptor.
Cell(Cambridge,Mass.), 142, 2010
|
|
6BVB
 
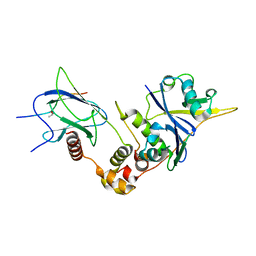 | | Crystal structure of HIF-2alpha-pVHL-elongin B-elongin C | | Descriptor: | Elongin-B, Elongin-C, Hypoxia-Inducible Factor 2 alpha, ... | | Authors: | Tarade, D, Ohh, M, Lee, J.E. | | Deposit date: | 2017-12-12 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | HIF-2 alpha-pVHL complex reveals broad genotype-phenotype correlations in HIF-2 alpha-driven disease.
Nat Commun, 9, 2018
|
|
1Q74
 
 | | The Crystal Structure of 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside Deacetylase (MshB) | | Descriptor: | 1D-myo-inositol 2-acetamido-2-deoxy-alpha-D-glucopyranoside Deacetylase (MshB), 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ZINC ION | | Authors: | Maynes, J.T, Garen, C, Cherney, M.M, Newton, G, Arad, D, Av-Gay, Y, Fahey, R.C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-08-15 | | Release date: | 2003-12-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of 1-D-myo-Inositol 2-Acetamido-2-deoxy-alpha-D-glucopyranoside Deacetylase (MshB) from Mycobacterium tuberculosis Reveals a Zinc Hydrolase with a Lactate Dehydrogenase Fold.
J.Biol.Chem., 278, 2003
|
|
3GOP
 
 | |
7UJV
 
 | | Structure of PHD2 in complex with HIF2a-CODD | | Descriptor: | Egl nine homolog 1, Endothelial PAS domain-containing protein 1, FE (III) ION, ... | | Authors: | Ferens, F.G, Tarade, D, Lee, J.E, Ohh, M. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deficiency in PHD2-mediated hydroxylation of HIF2 alpha underlies Pacak-Zhuang syndrome.
Commun Biol, 7, 2024
|
|
4Z8E
 
 | | TEAD DBD mutant -deltaL1 | | Descriptor: | Transcriptional enhancer factor TEF-1 | | Authors: | Lee, D.-S, Albarado, D.C, Vonrhein, C, Raman, C.S, Veeraraghavan, S. | | Deposit date: | 2015-04-08 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | A Potential Structural Switch for Regulating DNA-Binding by TEAD Transcription Factors.
J.Mol.Biol., 428, 2016
|
|
4WPC
 
 | |
4WPE
 
 | | Crystal Structure of Hof1p F-BAR domain | | Descriptor: | Cytokinesis protein 2 | | Authors: | Lemmon, M.A, Moravcevic, K. | | Deposit date: | 2014-10-17 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparison of Saccharomyces cerevisiae F-BAR Domain Structures Reveals a Conserved Inositol Phosphate Binding Site.
Structure, 23, 2015
|
|
5CUS
 
 | | Crystal Structure of sErbB3-Fab3379 Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab LC region of KTN3379, IgG H chain, ... | | Authors: | Lee, S, Schlessinger, J. | | Deposit date: | 2015-07-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inhibition of ErbB3 by a monoclonal antibody that locks the extracellular domain in an inactive configuration.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7LIR
 
 | | Structure of the invertebrate ALK GRD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALK tyrosine kinase receptor homolog scd-2, ... | | Authors: | Stayrook, S, Li, T, Klein, D.E. | | Deposit date: | 2021-01-27 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
7LS0
 
 | | Structure of the Human ALK GRD bound to AUG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALK tyrosine kinase receptor fused with ALK and LTK ligand 2, CITRIC ACID | | Authors: | Stayrook, S, Li, T, Klein, D.E. | | Deposit date: | 2021-02-17 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
7LRZ
 
 | | Structure of the Human ALK GRD | | Descriptor: | ALK tyrosine kinase receptor | | Authors: | Stayrook, S, Li, T, Klein, D.E. | | Deposit date: | 2021-02-17 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
7MK7
 
 | | Augmentor domain of augmentor-beta | | Descriptor: | ALK and LTK ligand 1,Maltodextrin-binding protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Krimmer, S.G, Reshetnyak, A.V, Puleo, D.E, Schlessinger, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42815185 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
