7XSA
 
 | |
7XSB
 
 | |
5UD8
 
 | | Crystal Structure of Mutant Ig-like Domain | | Descriptor: | Triggering receptor expressed on myeloid cells 2 | | Authors: | Sudom, A, Min, X, Wang, Z. | | Deposit date: | 2016-12-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for the loss-of-function effects of the Alzheimer's disease-associated R47H variant of the immune receptor TREM2.
J. Biol. Chem., 293, 2018
|
|
5UD7
 
 | | Crystal Structure of Wild-Type Ig-like Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IODIDE ION, SULFATE ION, ... | | Authors: | Sudom, A, Min, X, Wang, Z. | | Deposit date: | 2016-12-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.20002246 Å) | | Cite: | Molecular basis for the loss-of-function effects of the Alzheimer's disease-associated R47H variant of the immune receptor TREM2.
J. Biol. Chem., 293, 2018
|
|
5C6H
 
 | | Mcl-1 complexed with Mule | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Mule BH3 peptide from E3 ubiquitin-protein ligase HUWE1 | | Authors: | Song, T, Wang, Z, Ji, F, Chai, G, Liu, Y, Li, X, Li, Z, Fan, Y, Zhang, Z. | | Deposit date: | 2015-06-23 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Mcl-1 complexed with Mule at 2.05 Angstroms resolution
To Be Published
|
|
5UJV
 
 | | Crystal structure of FePYR1 in complex with abscisic acid | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, PYR1 | | Authors: | Ren, Z, Wang, Z, Zhou, X.E, Hong, Y, Cao, M, Chan, Z, Liu, X, Shi, H, Xu, H.E, Zhu, J.-K. | | Deposit date: | 2017-01-19 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure determination and activity manipulation of the turfgrass ABA receptor FePYR1.
Sci Rep, 7, 2017
|
|
5LZC
 
 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the codon reading state (CR) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZE
 
 | | Structure of the 70S ribosome with Sec-tRNASec in the classical pre-translocation state (C) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZB
 
 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the initial binding state (IB) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZD
 
 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the GTPase activated state (GA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
3FXV
 
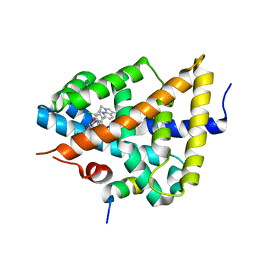 | | Identification of an N-oxide pyridine GW4064 analogue as a potent FXR agonist | | Descriptor: | 12-meric peptide from Nuclear receptor coactivator 1, 6-(4-{[3-(3,5-dichloropyridin-4-yl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}-2-methylphenyl)-1-methyl-1H-indole-3-carbox ylic acid, NR1H4 protein | | Authors: | Feng, S, Yang, M, He, Y, Chen, L, Zhang, Z, Wang, Z, Hong, D, Richter, H, Benson, G.M, Bleicher, K, Grether, U, Martin, R, Plancher, J.-M, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2009-01-21 | | Release date: | 2009-04-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Identification of an N-oxide pyridine GW4064 analog as a potent FXR agonist
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8JQ9
 
 | | Structure of Gabija GajA | | Descriptor: | Endonuclease GajA | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-06-13 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8JQC
 
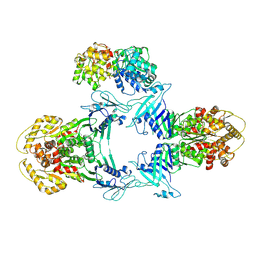 | | Structure of Gabija GajA-GajB 4:1 complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-06-13 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8JQB
 
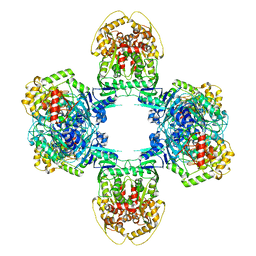 | | Structure of Gabija GajA-GajB 4:4 Complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-06-13 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
1N5U
 
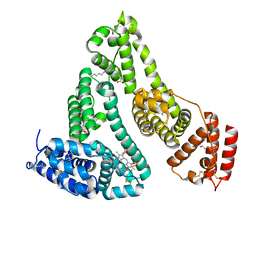 | | X-RAY STUDY OF HUMAN SERUM ALBUMIN COMPLEXED WITH HEME | | Descriptor: | MYRISTIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SERUM ALBUMIN | | Authors: | Wardell, M, Wang, Z, Ho, J.X, Robert, J, Ruker, F, Ruble, J, Carter, D.C. | | Deposit date: | 2002-11-07 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Atomic Structure of Human Methemalbumin at 1.9 A
Biochem.Biophys.Res.Commun., 291, 2002
|
|
2AX1
 
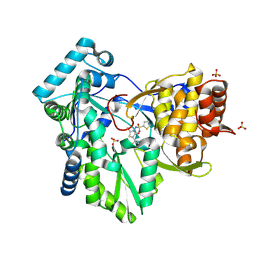 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5ee) | | Descriptor: | 5R-(3,4-DICHLOROPHENYLMETHYL)-3-(2-THIOPHENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | Authors: | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
1A40
 
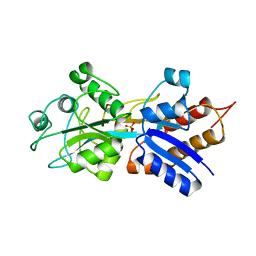 | | PHOSPHATE-BINDING PROTEIN WITH ALA 197 REPLACED WITH TRP | | Descriptor: | PHOSPHATE ION, PHOSPHATE-BINDING PERIPLASMIC PROTEIN PRECURSOR | | Authors: | Ledivina, P.S, Wang, Z, Tsai, A, Koehl, E, Quiocho, F.A. | | Deposit date: | 1998-02-10 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dominant role of local dipolar interactions in phosphate binding to a receptor cleft with an electronegative charge surface: equilibrium, kinetic, and crystallographic studies.
Protein Sci., 7, 1998
|
|
2AX0
 
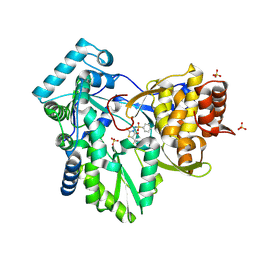 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5x) | | Descriptor: | 5R-(2E-METHYL-3-PHENYL-ALLYL)-3-(BENZENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | Authors: | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
2AWZ
 
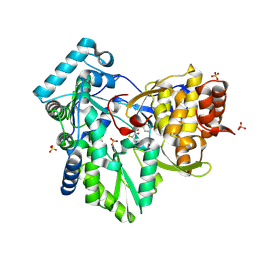 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5h) | | Descriptor: | 5R-(4-BROMOPHENYLMETHYL)-3-(BENZENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | Authors: | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
3BZU
 
 | | Crystal structure of human 11-beta-hydroxysteroid dehydrogenase(HSD1) in complex with NADP and thiazolone inhibitor | | Descriptor: | (5S)-2-{[(1S)-1-(2-fluorophenyl)ethyl]amino}-5-methyl-5-(trifluoromethyl)-1,3-thiazol-4(5H)-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Min, X, Sudom, A, Xu, H, Wang, Z, Walker, N.P. | | Deposit date: | 2008-01-18 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural characterization and pharmacodynamic effects of an orally active 11beta-hydroxysteroid dehydrogenase type 1 inhibitor.
Chem.Biol.Drug Des., 71, 2008
|
|
6PQP
 
 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the covalent agonist BITC | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-benzylthioformamide, ... | | Authors: | Suo, Y, Wang, Z, Zubcevic, L, Hsu, A.L, He, Q, Borgnia, M.J, Ji, R.-R, Lee, S.-Y. | | Deposit date: | 2019-07-09 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel.
Neuron, 105, 2020
|
|
6PQQ
 
 | | Cryo-EM structure of human TRPA1 C621S mutant in the apo state | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily A member 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Suo, Y, Wang, Z, Zubcevic, L, Hsu, A.L, He, Q, Borgnia, M.J, Ji, R.-R, Lee, S.-Y. | | Deposit date: | 2019-07-09 | | Release date: | 2020-01-08 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel.
Neuron, 105, 2020
|
|
6PQO
 
 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the covalent agonist JT010 | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloro-N-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-N-(3-methoxypropyl)acetamide, ... | | Authors: | Suo, Y, Wang, Z, Zubcevic, L, Hsu, A.L, He, Q, Borgnia, M.J, Ji, R.-R, Lee, S.-Y. | | Deposit date: | 2019-07-09 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel.
Neuron, 105, 2020
|
|
6Q04
 
 | | MERS-CoV S structure in complex with 5-N-acetyl neuraminic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6Q06
 
 | | MERS-CoV S structure in complex with 2,3-sialyl-N-acetyl-lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
