8P0M
 
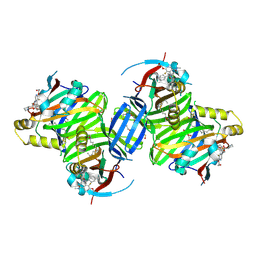 | | Crystal structure of TEAD3 in complex with IAG933 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 4-[(2~{S})-5-chloranyl-6-fluoranyl-2-phenyl-2-[(2~{S})-pyrrolidin-2-yl]-3~{H}-1-benzofuran-4-yl]-5-fluoranyl-6-(2-hydroxyethyloxy)-~{N}-methyl-pyridine-3-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Scheufler, C, Villard, F, Chau, S. | | Deposit date: | 2023-05-10 | | Release date: | 2024-04-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Direct and selective pharmacological disruption of the YAP-TEAD interface by IAG933 inhibits Hippo-dependent and RAS-MAPK-altered cancers.
Nat Cancer, 5, 2024
|
|
6TS0
 
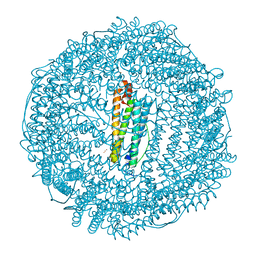 | | Crystal structure of human L ferritin (HuLf) triple variant E60A-E61A-E64A Fe(III)-loaded for 30 minutes | | Descriptor: | CADMIUM ION, FE (III) ION, Ferritin light chain, ... | | Authors: | Pozzi, C, Ciambellotti, S, Turano, P, Mangani, S. | | Deposit date: | 2019-12-19 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Iron Biomineral Growth from the Initial Nucleation Seed in L-Ferritin.
Chemistry, 26, 2020
|
|
6TSX
 
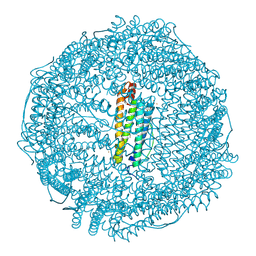 | | Crystal structure of horse L ferritin (HoLf) Fe(III)-loaded for 30 minutes | | Descriptor: | CADMIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Pozzi, C, Ciambellotti, S, Turano, P, Mangani, S. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.021 Å) | | Cite: | Iron Biomineral Growth from the Initial Nucleation Seed in L-Ferritin.
Chemistry, 26, 2020
|
|
6T61
 
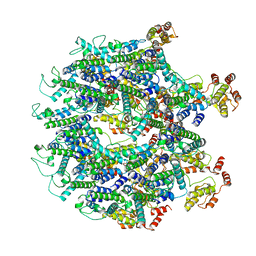 | | A model of the EIAV CA-SP hexamer (C2) from Gag-deltaMA tubes assembled at pH8 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
6T7H
 
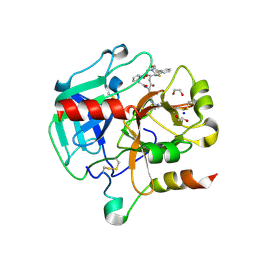 | | Crystal structure of Thrombin in complex with macrocycle N14-PR4-A | | Descriptor: | (14S,17R)-14-(3-carbamimidamidopropyl)-3-(furan-2-ylmethyl)-5,12,15-tris(oxidanylidene)-19-thia-3,6,13,16-tetrazatricyclo[19.4.0.0^{6,10}]pentacosa-1(25),7,9,21,23-pentaene-17-carboxamide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Angelini, A, Kumar, M.G, Heinis, C, Cendron, L. | | Deposit date: | 2019-10-22 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Macrocycle synthesis strategy based on step-wise "adding and reacting" three components enables screening of large combinatorial libraries.
Chem Sci, 11, 2020
|
|
6TNZ
 
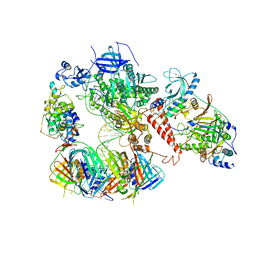 | | Human polymerase delta-FEN1-PCNA toolbelt | | Descriptor: | DNA polymerase delta catalytic subunit, DNA polymerase delta subunit 2, DNA polymerase delta subunit 3, ... | | Authors: | Lancey, C, Hamdan, S.M, De Biasio, A. | | Deposit date: | 2019-12-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structure of the processive human Pol delta holoenzyme.
Nat Commun, 11, 2020
|
|
6T7X
 
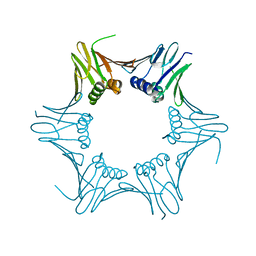 | | Crystal structure of PCNA from P. abyssi | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
6TQ6
 
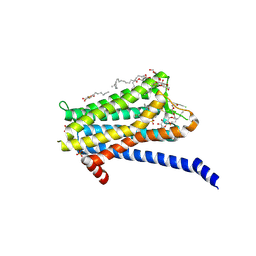 | | Crystal structure of the Orexin-1 receptor in complex with Compound 14 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-(5-methylsulfonylpyridin-3-yl)-1,1-bis(oxidanylidene)-4-[[2,4,6-tris(fluoranyl)phenyl]methyl]pyrido[2,3-e][1,2,4]thiadiazin-3-one, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.546 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6TGY
 
 | |
6TBX
 
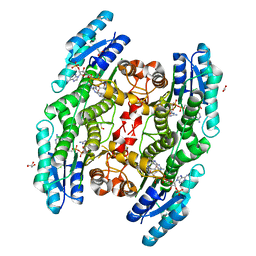 | | Trypanosoma brucei PTR1 (TbPTR1) in complex with a tricyclic-based inhibitor | | Descriptor: | 2,4-bis(azanyl)-9~{H}-pyrimido[4,5-b]indol-6-ol, ACETATE ION, GLYCEROL, ... | | Authors: | Landi, G, Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2019-11-04 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution crystal structure of Trypanosoma brucei pteridine reductase 1 in complex with an innovative tricyclic-based inhibitor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6TEQ
 
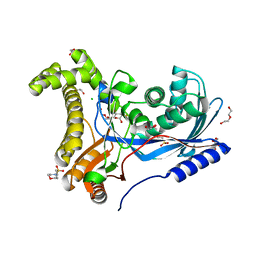 | | Crystal structure of a galactokinase from Bifidobacterium infantis in complex with 2-deoxy-2-fluoro-galactose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-alpha-D-galactopyranose, ... | | Authors: | Keenan, T, Parmeggiani, F, Fontenelle, C.Q, Malassis, J, Vendeville, J, Offen, W.A, Both, P, Huang, K, Marchesi, A, Heyam, A, Young, C, Charnock, S, Davies, G.J, Linclau, B, Flitsch, S.L, Fascione, M.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Profiling Substrate Promiscuity of Wild-Type Sugar Kinases for Multi-fluorinated Monosaccharides.
Cell Chem Biol, 27, 2020
|
|
6TUR
 
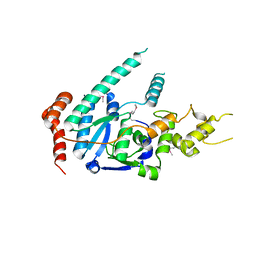 | | human XPG, Apo1 form | | Descriptor: | DNA repair protein complementing XP-G cells,DNA repair protein complementing XP-G cells | | Authors: | Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2020-01-08 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of human XPG, the xeroderma pigmentosum group G endonuclease, provides insight into nucleotide excision DNA repair.
Nucleic Acids Res., 48, 2020
|
|
6T2W
 
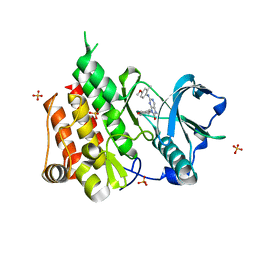 | | Crystal structure of the CSF1R kinase domain with a dihydropurinone inhibitor (compound 4) | | Descriptor: | 2-[(4-methoxy-2-methyl-phenyl)amino]-7-methyl-9-(4-oxidanylcyclohexyl)purin-8-one, Macrophage colony-stimulating factor 1 receptor, SULFATE ION | | Authors: | Schimpl, M, Goldberg, F.W, Finlay, M.R.V, Ting, A.K.T, Beattie, D, Lamont, G.M, Fallan, C, Wrigley, G.L, Howard, M.R, Williamson, B, Davies, B.R, Cadogan, E.B, Ramos-Montoya, A, Dean, E. | | Deposit date: | 2019-10-09 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Discovery of 7-Methyl-2-[(7-methyl[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8-one (AZD7648), a Potent and Selective DNA-Dependent Protein Kinase (DNA-PK) Inhibitor.
J.Med.Chem., 63, 2020
|
|
6THP
 
 | |
6T3M
 
 | | Thrombin in Complex with a D-Phe-Pro-p-phenol derivative | | Descriptor: | (2~{S})-1-[(2~{R})-2-azanyl-3-phenyl-propanoyl]-~{N}-[(4-hydroxyphenyl)methyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin variant-2, ... | | Authors: | Ngo, K, Collins, C, Heine, A, Klebe, G. | | Deposit date: | 2019-10-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Thrombin in Complex with a D-Phe-Pro-p-phenol derivative
To Be Published
|
|
6T63
 
 | | A model of the EIAV CA-SP hexamer (C2) from Gag-deltaMA tubes assembled at pH6 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
6T78
 
 | | Structure of human Sox11 transcription factor in complex with a short DNA fragment | | Descriptor: | DNA (5'-D(*AP*AP*CP*AP*AP*AP*AP*TP*AP*AP*AP*CP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*TP*GP*TP*TP*TP*AP*TP*TP*TP*TP*GP*TP*T)-3'), Transcription factor SOX-11 | | Authors: | Dodonova, S.O, Zhu, F, Dienemann, C, Taipale, J, Cramer, P. | | Deposit date: | 2019-10-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function.
Nature, 580, 2020
|
|
6T8N
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K3007 | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type I, DIMETHYL SULFOXIDE, ... | | Authors: | Adamson, R.J, Williams, E.P, Bonomo, S, Rankin, S, Bacos, D, Rae, A, Cramp, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-10-24 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K3007
To Be Published
|
|
6TCY
 
 | |
6TD4
 
 | | IRF4 DNA-binding domain surface entropy mutant apo structure | | Descriptor: | CHLORIDE ION, Interferon regulatory factor 4 | | Authors: | Tucker, J.A, Martin, M.P, Wang, L.Z, Jennings, C, Heath, R. | | Deposit date: | 2019-11-07 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Cancer-associated mutations in the IRF4 DNA-binding domain confer no disadvantage in DNA-binding affinity and may increase transcriptional activity
To Be Published
|
|
6TFA
 
 | |
6TBM
 
 | | Structure of SAGA bound to TBP, including Spt8 and DUB | | Descriptor: | Polyubiquitin-B, SAGA-associated factor 11, Spt20, ... | | Authors: | Papai, G, Frechard, A, Kolesnikova, O, Crucifix, C, Schultz, P, Ben-Shem, A. | | Deposit date: | 2019-11-01 | | Release date: | 2020-02-12 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structure of SAGA and mechanism of TBP deposition on gene promoters.
Nature, 577, 2020
|
|
6TDA
 
 | | Structure of SWI/SNF chromatin remodeler RSC bound to a nucleosome | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC58, ... | | Authors: | Wagner, F.R, Dienemann, C, Wang, H, Stuetzer, A, Tegunov, D, Urlaub, H, Cramer, P. | | Deposit date: | 2019-11-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome.
Nature, 579, 2020
|
|
9FO2
 
 | | Coxsackievirus A9 bound with compound 15 (CL278) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Plavec, Z, Butcher, S.J, Mitchell, C, Buckner, C. | | Deposit date: | 2024-06-11 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | SAR Analysis of Novel Coxsackie virus A9 Capsid Binders.
J.Med.Chem., 67, 2024
|
|
6TRD
 
 | | Cryo- EM structure of the Thermosynechococcus elongatus photosystem I in the presence of cytochrome c6 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Koelsch, A, Radon, C, Baumert, A, Buerger, J, Mielke, T, Lisdat, F, Zouni, A, Wendler, P. | | Deposit date: | 2019-12-18 | | Release date: | 2020-09-16 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Current limits of structural biology: The transient interaction between cytochrome c6 and photosystem I
Curr.Opin.Struct.Biol., 2, 2020
|
|
