6TG9
 
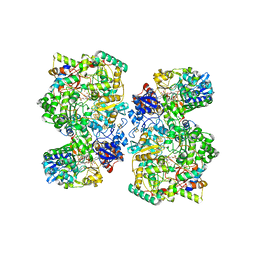 | | Cryo-EM Structure of NADH reduced form of NAD+-dependent Formate Dehydrogenase from Rhodobacter capsulatus | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wendler, P, Radon, C, Mittelstaedt, G. | | Deposit date: | 2019-11-15 | | Release date: | 2020-04-22 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structures reveal intricate Fe-S cluster arrangement and charging in Rhodobacter capsulatus formate dehydrogenase.
Nat Commun, 11, 2020
|
|
6TGA
 
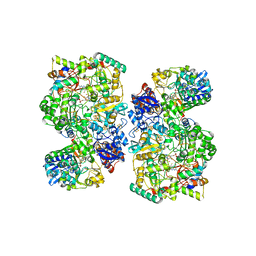 | | Cryo-EM Structure of as isolated form of NAD+-dependent Formate Dehydrogenase from Rhodobacter capsulatus | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wendler, P, Radon, C, Mittelstaedt, G. | | Deposit date: | 2019-11-15 | | Release date: | 2020-04-22 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Cryo-EM structures reveal intricate Fe-S cluster arrangement and charging in Rhodobacter capsulatus formate dehydrogenase.
Nat Commun, 11, 2020
|
|
3OC3
 
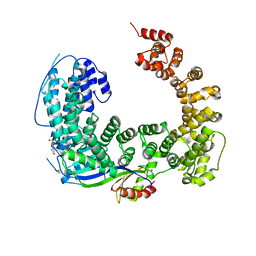 | | Crystal structure of the Mot1 N-terminal domain in complex with TBP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HELICASE MOT1, TRANSCRIPTION INITIATION FACTOR TFIID (TFIID-1) | | Authors: | Wollmann, P, Cui, S, Viswanathan, R, Berninghausen, O, Wells, M.N, Moldt, M, Witte, G, Butryn, A, Wendler, P, Beckmann, R, Auble, D.T, Hopfner, K.-P. | | Deposit date: | 2010-08-09 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP.
Nature, 475, 2011
|
|
5NV3
 
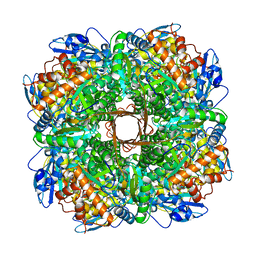 | | Structure of Rubisco from Rhodobacter sphaeroides in complex with CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Bracher, A, Milicic, G, Ciniawsky, S, Wendler, P, Hayer-Hartl, M, Hartl, F.U. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-26 | | Last modified: | 2017-09-20 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Mechanism of Enzyme Repair by the AAA(+) Chaperone Rubisco Activase.
Mol. Cell, 67, 2017
|
|
3ZUH
 
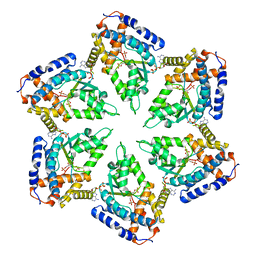 | | Negative stain EM Map of the AAA protein CbbX, a red-type Rubisco activase from R. sphaeroides | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PROTEIN CBBX, RIBULOSE-1,5-DIPHOSPHATE | | Authors: | Mueller-Cajar, O, Stotz, M, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-09 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | Structure and Function of the Aaa+ Protein Cbbx, a Red-Type Rubisco Activase.
Nature, 479, 2011
|
|
3ZW6
 
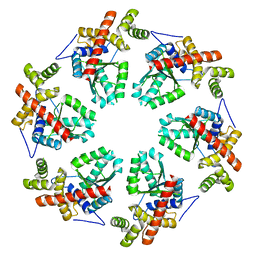 | | MODEL OF HEXAMERIC AAA DOMAIN ARRANGEMENT OF GREEN-TYPE RUBISCO ACTIVASE FROM TOBACCO. | | Descriptor: | RIBULOSE BISPHOSPHATE CARBOXYLASE/OXYGENASE ACTIVASE 1, CHLOROPLASTIC | | Authors: | Stotz, M, Mueller-Cajar, O, Ciniawsky, S, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-09 | | Last modified: | 2019-10-30 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structure of Green-Type Rubisco Activase from Tobacco
Nat.Struct.Mol.Biol., 18, 2011
|
|
4WT3
 
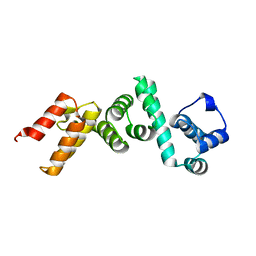 | | The N-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana | | Descriptor: | Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4WT4
 
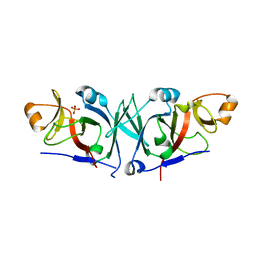 | | The C-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana, crystal form I | | Descriptor: | PHOSPHATE ION, Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4WT5
 
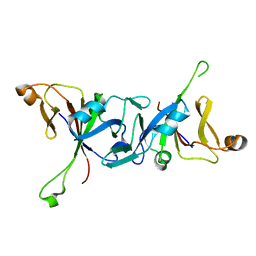 | | The C-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana, crystal form II | | Descriptor: | Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3T15
 
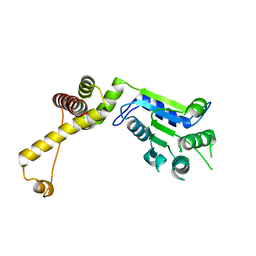 | | Structure of green-type Rubisco activase from tobacco | | Descriptor: | Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplastic | | Authors: | Stotz, M, Wendler, P, Mueller-Cajar, O, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2011-07-21 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of green-type Rubisco activase from tobacco.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3SYL
 
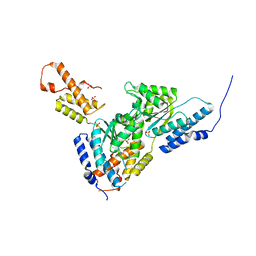 | | Crystal structure of the AAA+ protein CbbX, native structure | | Descriptor: | Protein CbbX, SULFATE ION | | Authors: | Mueller-Cajar, O, Stotz, M, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2011-07-18 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase.
Nature, 479, 2011
|
|
3SYK
 
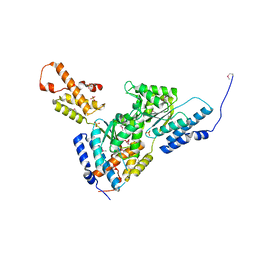 | | Crystal structure of the AAA+ protein CbbX, selenomethionine structure | | Descriptor: | Protein CbbX, SULFATE ION | | Authors: | Mueller-Cajar, O, Stotz, M, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2011-07-18 | | Release date: | 2011-11-09 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase.
Nature, 479, 2011
|
|
6TRC
 
 | | Cryo- EM structure of the Thermosynechococcus elongatus photosystem I in the presence of cytochrome c6 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Koelsch, A, Radon, C, Baumert, A, Buerger, J, Mielke, T, Lisdat, F, Zouni, A, Wendler, P. | | Deposit date: | 2019-12-18 | | Release date: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Current limits of structural biology: The transient interaction between cytochrome c6 and photosystem I
Curr.Opin.Struct.Biol., 2, 2020
|
|
6TRA
 
 | | Cryo- EM structure of the Thermosynechococcus elongatus photosystem I in the presence of cytochrome c6 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Koelsch, A, Radon, C, Baumert, A, Buerger, J, Miehlke, T, Lisdat, F, Zouni, A, Wendler, P. | | Deposit date: | 2019-12-18 | | Release date: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Current limits of structural biology: The transient interaction between cytochrome c6 and photosystem I
Curr.Opin.Struct.Biol., 2, 2020
|
|
6TRD
 
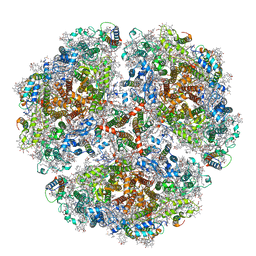 | | Cryo- EM structure of the Thermosynechococcus elongatus photosystem I in the presence of cytochrome c6 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Koelsch, A, Radon, C, Baumert, A, Buerger, J, Mielke, T, Lisdat, F, Zouni, A, Wendler, P. | | Deposit date: | 2019-12-18 | | Release date: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Current limits of structural biology: The transient interaction between cytochrome c6 and photosystem I
Curr.Opin.Struct.Biol., 2, 2020
|
|
4D2Q
 
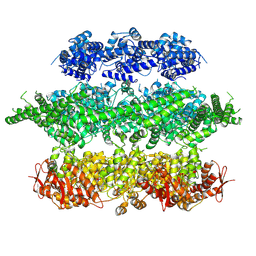 | | Negative-stain electron microscopy of E. coli ClpB mutant E432A (BAP form bound to ClpP) | | Descriptor: | CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-12 | | Release date: | 2014-06-04 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
4D2X
 
 | | Negative-stain electron microscopy of E. coli ClpB of Y503D hyperactive mutant (BAP form bound to ClpP) | | Descriptor: | CHAPERONE PROTEIN CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-04 | | Last modified: | 2019-01-23 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
4D2U
 
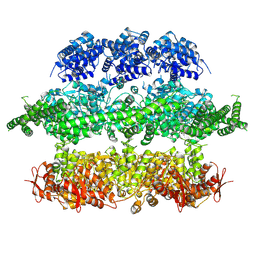 | | Negative-stain electron microscopy of E. coli ClpB (BAP form bound to ClpP) | | Descriptor: | CHAPERONE PROTEIN CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-04 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
