6YVE
 
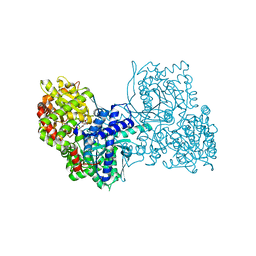 | | Glycogen phosphorylase b in complex with pelargonidin 3-O-beta-D-glucoside | | Descriptor: | DIMETHYL SULFOXIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Drakou, C.E, Gardeli, C, Tsialtas, I, Alexopoulos, S, Mallouchos, A, Koulas, S, Tsagkarakou, A, Asimakopoulos, D, Leonidas, D.D, Psarra, A.M, Skamnaki, V.T. | | Deposit date: | 2020-04-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Affinity Crystallography Reveals Binding of Pomegranate Juice Anthocyanins at the Inhibitor Site of Glycogen Phosphorylase: The Contribution of a Sugar Moiety to Potency and Its Implications to the Binding Mode.
J.Agric.Food Chem., 68, 2020
|
|
4UI2
 
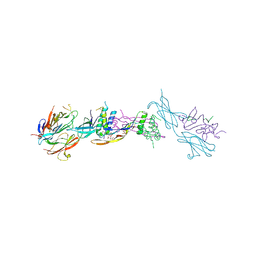 | | Crystal structure of the ternary RGMB-BMP2-NEO1 complex | | Descriptor: | ACETATE ION, BONE MORPHOGENETIC PROTEIN 2, BMP2, ... | | Authors: | Healey, E.G, Bishop, B, Elegheert, J, Bell, C.H, Padilla-Parra, S, Siebold, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-06 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Repulsive Guidance Molecule is a Structural Bridge between Neogenin and Bone Morphogenetic Protein.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6YYT
 
 | | Structure of replicating SARS-CoV-2 polymerase | | Descriptor: | RNA product, ZINC ION, nsp12, ... | | Authors: | Hillen, H.S, Kokic, G, Farnung, L, Dienemann, C, Tegunov, D, Cramer, P. | | Deposit date: | 2020-05-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of replicating SARS-CoV-2 polymerase.
Nature, 584, 2020
|
|
4UOV
 
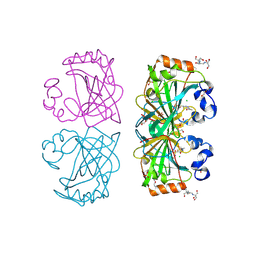 | | The structure of a tetrameric alpha-carbonic anhydrase from Thermovibrio ammonificans reveals a core formed around intermolecular disulfides, which contribute to its thermostability. | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | James, P, Isupov, M, Sayer, C, Berg, S, Lioliou, M, Kotlar, H, Littlechild, J. | | Deposit date: | 2014-06-10 | | Release date: | 2014-10-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of a Tetrameric [Alpha]-Carbonic Anhydrase from Thermovibrio Ammonificans Reveals a Core Formed Around Intermolecular Disulfides that Contribute to its Thermostability
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6YT0
 
 | |
4UIY
 
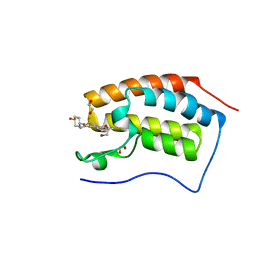 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH N-(1,1-dioxo-1-thian-4-yl)- 5-methyl-4-oxo-7-3-(trifluoromethyl)phenyl-4H,5H-thieno-3,2-c- pyridine-2-carboximidamide | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN-CONTAINING PROTEIN 4, N-[1,1-bis(oxidanylidene)thian-4-yl]-5-methyl-4-oxidanylidene-7-[3-(trifluoromethyl)phenyl]thieno[3,2-c]pyridine-2-carboximidamide | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
7FEE
 
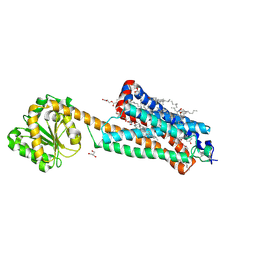 | | Crystal structure of the allosteric modulator ZCZ011 binding to CP55940-bound cannabinoid receptor 1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, 6-methyl-3-[(1S)-2-nitro-1-thiophen-2-yl-ethyl]-2-phenyl-1H-indole, ... | | Authors: | Wang, X, Zhao, C, Shao, Z. | | Deposit date: | 2021-07-19 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular mechanism of allosteric modulation for the cannabinoid receptor CB1.
Nat.Chem.Biol., 18, 2022
|
|
4UCI
 
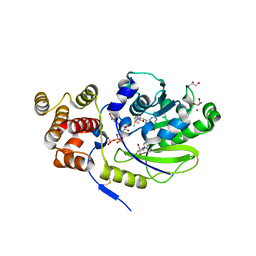 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | ADENOSINE, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-03 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
6YV7
 
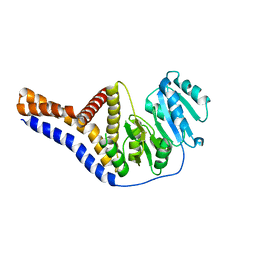 | | Mannosyltransferase PcManGT from Pyrobaculum calidifontis | | Descriptor: | Glycosyl transferase, family 2 | | Authors: | Divne, C, Rosaria, G. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Transmembrane Crenarchaeal Mannosyltransferase Is Involved in N-Glycan Biosynthesis and Displays an Unexpected Minimal Cellulose-Synthase-like Fold.
J.Mol.Biol., 432, 2020
|
|
6YVV
 
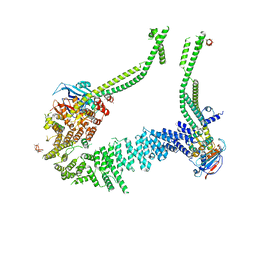 | | Condensin complex from S.cerevisiae ATP-free apo bridged state | | Descriptor: | Condensin complex subunit 1,Ycs4, Condensin complex subunit 2,Brn1, Structural maintenance of chromosomes protein 2,Structural maintenance of chromosomes protein 2, ... | | Authors: | Lee, B.-G, Cawood, C, Gutierrez-Escribano, P, Nakane, T, Merkel, F, Hassler, M, Haering, C.H, Aragon, L, Lowe, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4UI0
 
 | | Crystal structure of the human RGMB-BMP2 complex, crystal form 2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BONE MORPHOGENETIC PROTEIN 2, CHLORIDE ION, ... | | Authors: | Healey, E.G, Bishop, B, Elegheert, J, Bell, C.H, Padilla-Parra, S, Siebold, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Repulsive Guidance Molecule is a Structural Bridge between Neogenin and Bone Morphogenetic Protein.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6YWD
 
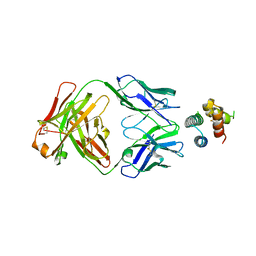 | | De novo designed protein 4H_01 in complex with Mota antibody | | Descriptor: | Antibody Mota, Heavy Chain, Light Chain, ... | | Authors: | Yang, C, Sesterhenn, F, Pojer, F, Correia, B.E. | | Deposit date: | 2020-04-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Bottom-up de novo design of functional proteins with complex structural features.
Nat.Chem.Biol., 17, 2021
|
|
4UCV
 
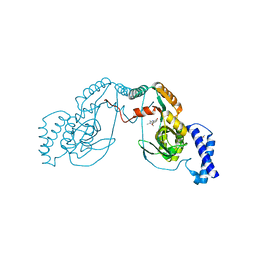 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-methoxy-2,3-dimethylquinoxalin-5-ol, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
6YTJ
 
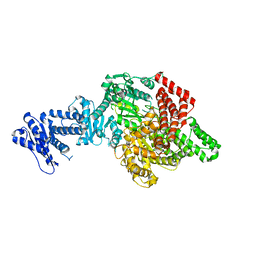 | |
4UHN
 
 | | Characterization of a Novel Transaminase from Pseudomonas sp. Strain AAC | | Descriptor: | ACETIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wilding, M, Peat, T.S, Newman, J, Scott, C. | | Deposit date: | 2015-03-25 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A Beta-Alanine Catabolism Pathway Containing a Highly Promiscuous Omega-Transaminase in the 12-Aminododecanate-Degrading Pseudomonas Sp. Strain Aac.
Appl.Environ.Microbiol., 82, 2016
|
|
4UHX
 
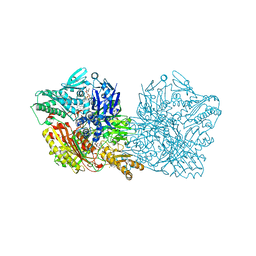 | | Human aldehyde oxidase in complex with phthalazine and thioridazine | | Descriptor: | 10-{2-[(2R)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, 10-{2-[(2S)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, ALDEHYDE OXIDASE, ... | | Authors: | Coelho, C, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2015-03-26 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights Into Xenobiotic and Inhibitor Binding to Human Aldehyde Oxidase
Nat.Chem.Biol., 11, 2015
|
|
6LCU
 
 | |
4UIS
 
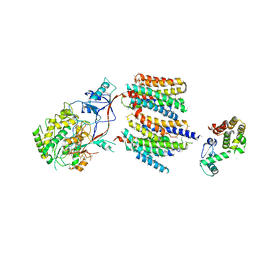 | | The cryoEM structure of human gamma-Secretase complex | | Descriptor: | GAMMA-SECRETASE, LYSOZYME | | Authors: | Sun, L, Zhao, L, Yang, G, Yan, C, Zhou, R, Zhou, X, Xie, T, Zhao, Y, Wu, S, Li, X, Shi, Y. | | Deposit date: | 2015-04-03 | | Release date: | 2015-06-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis of Human Gamma-Secretase Assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4UIW
 
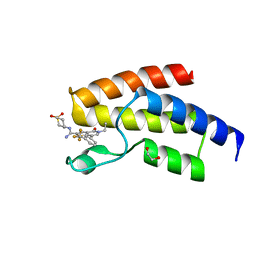 | | BROMODOMAIN OF HUMAN BRD9 WITH N-(1,1-dioxo-1-thian-4-yl)-5-ethyl-4- oxo-7-3-(trifluoromethyl)phenyl-4H,5H-thieno-3,2-c-pyridine-2- carboximidamide | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN-CONTAINING PROTEIN 9, N'-[1,1-bis(oxidanylidene)thian-4-yl]-5-ethyl-4-oxidanylidene-7-[3-(trifluoromethyl)phenyl]thieno[3,2-c]pyridine-2-carboximidamide | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
6YU5
 
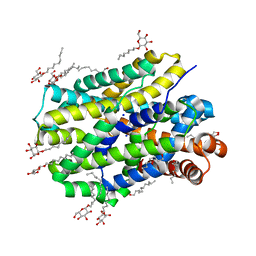 | | Crystal structure of MhsT in complex with L-valine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6LEN
 
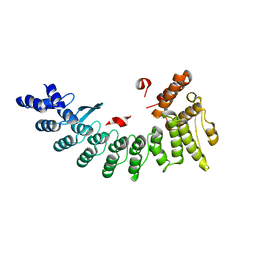 | | Structure of NS11 bound FEM1C | | Descriptor: | Protein fem-1 homolog C,NS11 peptide | | Authors: | Chen, x, Liao, S, Xu, C. | | Deposit date: | 2019-11-25 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
4UN2
 
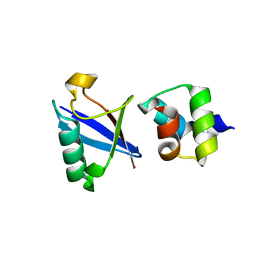 | | Crystal structure of the UBA domain of Dsk2 in complex with Ubiquitin | | Descriptor: | UBIQUITIN, UBIQUITIN DOMAIN-CONTAINING PROTEIN DSK2 | | Authors: | Michielssens, S, Peters, J.H, Ban, D, Pratihar, S, Seeliger, D, Sharma, M, Giller, K, Sabo, T.M, Becker, S, Lee, D, Griesinger, C, de Groot, B.L. | | Deposit date: | 2014-05-23 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A Designed Conformational Shift to Control Protein Binding Specificity.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6KY7
 
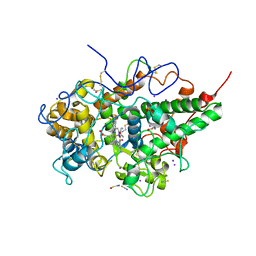 | | Crystal structure of yak lactoperoxidase at 2.27 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, P.K, Viswanathan, V, Sharma, P, Rani, C, Sharma, S, Singh, T.P. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of yak lactoperoxidase at 2.27 A resolution
To Be Published
|
|
6Z30
 
 | | Human cation-independent mannose 6-phosphate/ IGF2 receptor domains 9-10 | | Descriptor: | Cation-independent mannose-6-phosphate receptor, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bochel, A.J, Williams, C, Crump, M.P. | | Deposit date: | 2020-05-19 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7-11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9.
Structure, 28, 2020
|
|
6Z36
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2118 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-(4-piperidin-4-ylphenyl)-5-(3,4,5-trimethoxyphenyl)pyridine, AMMONIUM ION, ... | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-05-19 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2118
To Be Published
|
|
