5QIN
 
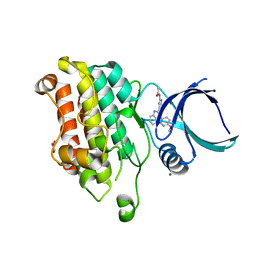 | |
5QIM
 
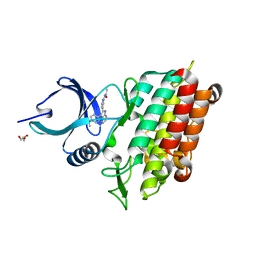 | |
5QIL
 
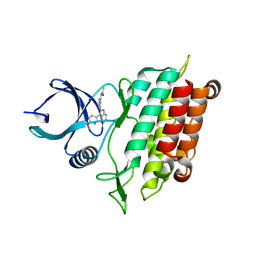 | |
5QIK
 
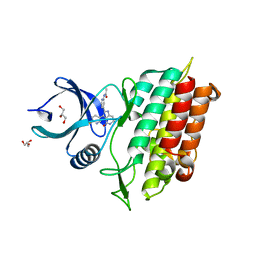 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) IN COMPLEX WITH N-{4-[3-(6-fluoropyridin-3-yl)-4-oxo-4,5,6,7-tetrahydro-1H-pyrrolo[3,2-c]pyridin-2-yl]pyridin-2-yl}acetamide | | Descriptor: | GLYCEROL, N-{4-[3-(6-fluoropyridin-3-yl)-4-oxo-4,5,6,7-tetrahydro-1H-pyrrolo[3,2-c]pyridin-2-yl]pyridin-2-yl}acetamide, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2018-08-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of 4-Azaindole Inhibitors of TGF beta RI as Immuno-oncology Agents.
ACS Med Chem Lett, 9, 2018
|
|
6Q1F
 
 | | Atomic structure of the Human Herpesvirus 6B Capsid and Capsid-Associated Tegument Complexes | | Descriptor: | Large structural phosphoprotein, Major capsid protein, Small capsomere-interacting protein, ... | | Authors: | Zhang, Y.B, Liu, W, Li, Z.H, Kumar, V, Alvarez-Cabrera, A.L, Leibovitch, E, Cui, Y.X, Mei, Y, Bi, G.Q, Jacobson, S, Zhou, Z.H. | | Deposit date: | 2019-08-03 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Atomic structure of the human herpesvirus 6B capsid and capsid-associated tegument complexes.
Nat Commun, 10, 2019
|
|
3CC0
 
 | |
3CBX
 
 | |
3CBY
 
 | |
3CBZ
 
 | |
7OS4
 
 | | Crystal structure of mouse CARM1 in complex with histone H3_13-31 K18 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, Histone H3.1, Histone-arginine methyltransferase CARM1 | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-06-07 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural Studies Provide New Insights into the Role of Lysine Acetylation on Substrate Recognition by CARM1 and Inform the Design of Potent Peptidomimetic Inhibitors.
Chembiochem, 22, 2021
|
|
7OKP
 
 | | Crystal structure of mouse CARM1 in complex with histone H3_13-22 K18 acetylated | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, Histone H3.3, Histone-arginine methyltransferase CARM1, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-05-18 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies Provide New Insights into the Role of Lysine Acetylation on Substrate Recognition by CARM1 and Inform the Design of Potent Peptidomimetic Inhibitors.
Chembiochem, 22, 2021
|
|
8CZ9
 
 | |
5E1R
 
 | |
6XRQ
 
 | | Structural descriptions of ligand interactions to DNA and RNA quadruplexes folded from the non-coding region of Pseudorabies virus | | Descriptor: | 2,7-bis[3-(morpholin-4-yl)propyl]-4,9-bis{[3-(morpholin-4-yl)propyl]amino}benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetrone, POTASSIUM ION, RNA (5' GP*GP*CP*UP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A-3') | | Authors: | Zhang, Y.S, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural descriptions of ligand interactions to RNA quadruplexes folded from the non-coding region of Pseudorabies virus.
Biochimie, 2024
|
|
6GFM
 
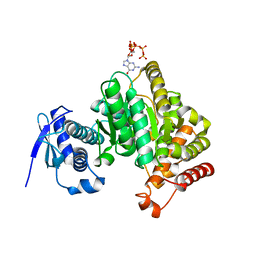 | | Crystal structure of the Escherichia coli nucleosidase PpnN (pppGpp-form) | | Descriptor: | Pyrimidine/purine nucleotide 5'-monophosphate nucleosidase, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Zhang, Y, Baerentsen, R.L, Gerdes, K, Brodersen, D.E. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | (p)ppGpp Regulates a Bacterial Nucleosidase by an Allosteric Two-Domain Switch.
Mol.Cell, 74, 2019
|
|
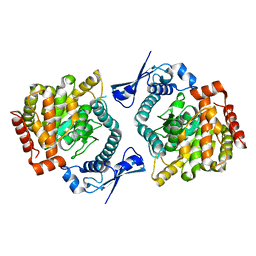
6GFL
 
 | |
4I6P
 
 | | Crystal structure of Par3-NTD domain | | Descriptor: | Partitioning defective 3 homolog | | Authors: | Wang, W, Gao, F, Gong, W, Sun, F, Feng, W. | | Deposit date: | 2012-11-29 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the intrinsic self-assembly of par-3 N-terminal domain.
Structure, 21, 2013
|
|
4QQG
 
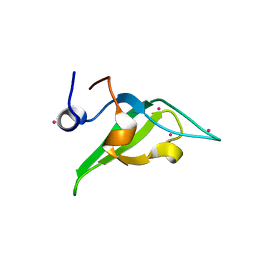 | | Crystal structure of an N-terminal HTATIP fragment | | Descriptor: | Histone acetyltransferase KAT5, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Wernimont, A.K, Dobrovetsky, E, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and histone binding studies of the chromo barrel domain of TIP60.
FEBS Lett., 592, 2018
|
|
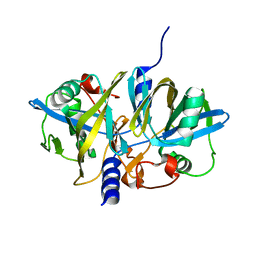
3NFW
 
 | |
4RZZ
 
 | |
4S00
 
 | |
4RZY
 
 | |
4S01
 
 | |
4Z4P
 
 | | Structure of the MLL4 SET Domain | | Descriptor: | Histone-lysine N-methyltransferase 2D, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Zhang, Z, Mittal, A, Reid, J, Reich, S, Gamblin, S.J, Wilson, J.R. | | Deposit date: | 2015-04-02 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolving Catalytic Properties of the MLL Family SET Domain.
Structure, 23, 2015
|
|
8JD9
 
 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Sodium/hydrogen exchanger 7 | | Authors: | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
