5ADY
 
 | | Cryo-EM structures of the 50S ribosome subunit bound with HflX | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L10, ... | | Authors: | Zhang, Y, Mandava, C.S, Cao, W, Li, X, Zhang, D, Li, N, Zhang, Y, Zhang, X, Qin, Y, Mi, K, Lei, J, Sanyal, S, Gao, N. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-14 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Hflx is a Ribosome Splitting Factor Rescuing Stalled Ribosomes Under Stress Conditions
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XV2
 
 | |
4XV3
 
 | | B-Raf Kinase V600E oncogenic mutant in complex with PLX7922 | | Descriptor: | N'-{3-[5-(2-aminopyrimidin-4-yl)-2-tert-butyl-1,3-thiazol-4-yl]-2-fluorophenyl}-N-ethyl-N-methylsulfuric diamide, Serine/threonine-protein kinase B-raf | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RAF inhibitors that evade paradoxical MAPK pathway activation.
Nature, 526, 2015
|
|
4XV9
 
 | | B-Raf Kinase domain in complex with PLX5568 | | Descriptor: | N-{3-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]-2,4-difluorophenyl}-4-(trifluoromethyl)benzenesulfonamide, SULFATE ION, Serine/threonine-protein kinase B-raf | | Authors: | zhang, Y, zhang, c, wang, w. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RAF inhibitors that evade paradoxical MAPK pathway activation.
Nature, 526, 2015
|
|
4XV1
 
 | | B-Raf Kinase V600E oncogenic mutant in complex with PLX7904 | | Descriptor: | N'-(3-{[5-(2-cyclopropylpyrimidin-5-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl]carbonyl}-2,4-difluorophenyl)-N-ethyl-N-methylsulfuric diamide, Serine/threonine-protein kinase B-raf | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | RAF inhibitors that evade paradoxical MAPK pathway activation.
Nature, 526, 2015
|
|
5LQ6
 
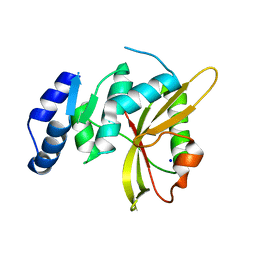 | | Salmonella effector SpvD - R161 variant | | Descriptor: | SODIUM ION, Virulence protein vsdE | | Authors: | Zhang, Y, Grabe, G.J, Rolhion, N, Yang, Y, Holden, D.W, Hare, S.A. | | Deposit date: | 2016-08-16 | | Release date: | 2016-11-02 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The Salmonella Effector SpvD Is a Cysteine Hydrolase with a Serovar-specific Polymorphism Influencing Catalytic Activity, Suppression of Immune Responses, and Bacterial Virulence.
J. Biol. Chem., 291, 2016
|
|
8XRY
 
 | |
8XS5
 
 | |
8XVX
 
 | |
8XW2
 
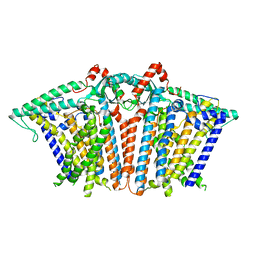 | |
8XVY
 
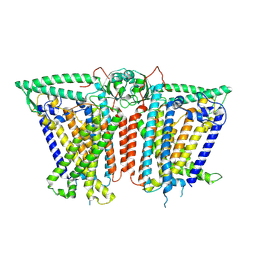 | |
8XW3
 
 | |
8XAJ
 
 | | Cryo-EM structure of OSCA1.2-liposome-inside-in open state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Calcium permeable stress-gated cation channel 1 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 2024
|
|
8XS4
 
 | |
8XNG
 
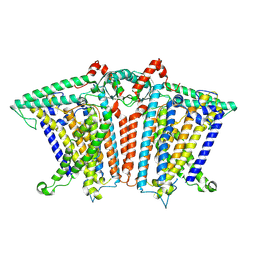 | |
8XVZ
 
 | |
8XS0
 
 | |
8XW0
 
 | | Cryo-EM structure of OSCA3.1-GDN state | | Descriptor: | CSC1-like protein ERD4, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 2024
|
|
8XW1
 
 | | Cryo-EM structure of OSCA1.2-V335W-DDM state | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.49 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 2024
|
|
8XW4
 
 | |
6XNZ
 
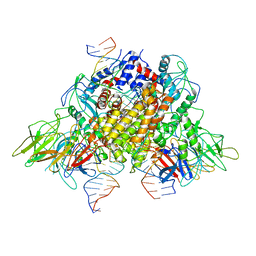 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Target Capture Complex | | Descriptor: | 12RSS integration strand (34-mer), 12RSS non-integration strand (34-mer), 23RSS integration strand (45-mer), ... | | Authors: | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
6XNY
 
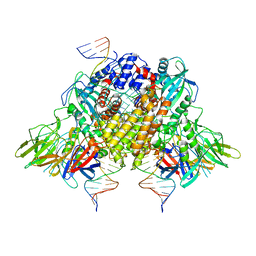 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Strand Transfer Complex (Paired-Form) | | Descriptor: | 12RSS integration strand (55-mer), 12RSS signal DNA top strand (34-mer), 23RSS integration strand (66-mer), ... | | Authors: | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
6Y5B
 
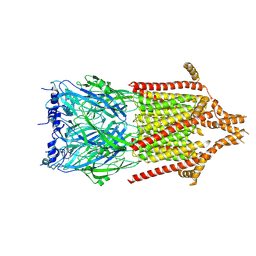 | | 5-HT3A receptor in Salipro (apo, asymmetric) | | Descriptor: | 5-hydroxytryptamine receptor 3A | | Authors: | Zhang, Y, Dijkman, P.M, Zou, R, Zandl-Lang, M, Sanchez, R.M, Eckhardt-Strelau, L, Koefeler, H, Vogel, H, Yuan, S, Kudryashev, M. | | Deposit date: | 2020-02-25 | | Release date: | 2020-12-23 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Asymmetric opening of the homopentameric 5-HT 3A serotonin receptor in lipid bilayers.
Nat Commun, 12, 2021
|
|
4R7H
 
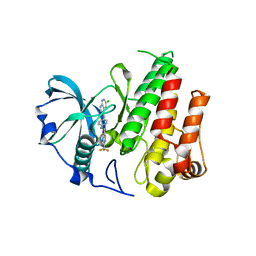 | | Crystal structure of FMS KINASE domain with a small molecular inhibitor, PLX3397 | | Descriptor: | 5-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]-N-{[6-(trifluoromethyl)pyridin-3-yl]methyl}pyridin-2-amine, Macrophage colony-stimulating factor 1 receptor | | Authors: | Zhang, Y, Zhang, K, Zhang, C. | | Deposit date: | 2014-08-27 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8001 Å) | | Cite: | Structure-Guided Blockade of CSF1R Kinase in Tenosynovial Giant-Cell Tumor.
N Engl J Med, 373, 2015
|
|
6Y59
 
 | | 5-HT3A receptor in Salipro (apo, C5 symmetric) | | Descriptor: | 5-hydroxytryptamine receptor 3A | | Authors: | Zhang, Y, Dijkman, P.M, Zou, R, Zandl-Lang, M, Sanchez, R.M, Eckhardt-Strelau, L, Koefeler, H, Vogel, H, Yuan, S, Kudryashev, M. | | Deposit date: | 2020-02-25 | | Release date: | 2020-12-23 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Asymmetric opening of the homopentameric 5-HT 3A serotonin receptor in lipid bilayers.
Nat Commun, 12, 2021
|
|
