3OUB
 
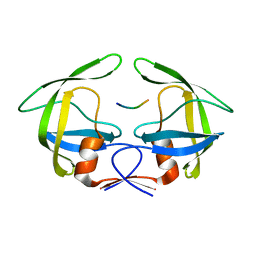 | | MDR769 HIV-1 protease complexed with NC/p1 hepta-peptide | | Descriptor: | MDR HIV-1 protease, NC/p1 substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3D7A
 
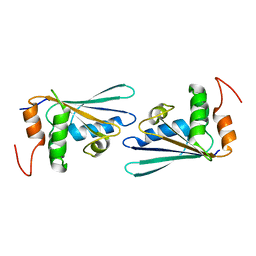 | |
2F0X
 
 | | Crystal structure and function of human thioesterase superfamily member 2(THEM2) | | Descriptor: | SULFATE ION, Thioesterase superfamily member 2 | | Authors: | Cheng, Z, Song, F, Shan, X, Wang, Y, Wei, Z, Gong, W. | | Deposit date: | 2005-11-14 | | Release date: | 2006-10-10 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human thioesterase superfamily member 2
Biochem.Biophys.Res.Commun., 349, 2006
|
|
3OUD
 
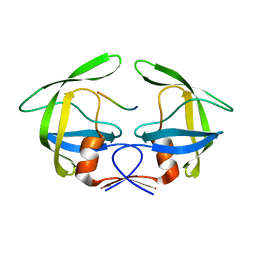 | | MDR769 HIV-1 protease complexed with CA/p2 hepta-peptide | | Descriptor: | CA/p2 substrate peptide, MDR HIV-1 protease | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3P8E
 
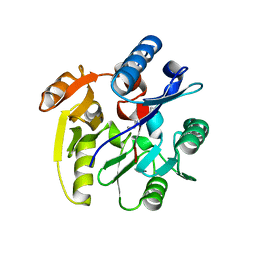 | | Crystal structure of human DIMETHYLARGININE DIMETHYLAMINOHYDROLASE-1 (DDAH-1) covalently bound with N5-(1-iminopentyl)-L-ornithine | | Descriptor: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, N~5~-[(1S)-1-aminopentyl]-L-ornithine | | Authors: | Lluis, M, Wang, Y, Monzingo, A.F, Fast, W, Robertus, J.D. | | Deposit date: | 2010-10-13 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4946 Å) | | Cite: | Characterization of C-Alkyl Amidines as Bioavailable Covalent Reversible Inhibitors of Human DDAH-1.
Chemmedchem, 6, 2011
|
|
3OXZ
 
 | | Crystal structure of ABL kinase domain bound with a DFG-out inhibitor AP24534 | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Tyrosine-protein kinase ABL1 | | Authors: | Zhou, T, Huang, W.S, Wang, Y, Thomas, M, Keats, J, Xu, Q, Rivera, V, Shakespeare, W.C, Clackson, T, Dalgarno, D.C, Zhu, X. | | Deposit date: | 2010-09-22 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib (AP24534): Lessons for Overcoming Kinase Inhibitor Resistance.
Chem.Biol.Drug Des., 77, 2011
|
|
3PP8
 
 | | 2.1 Angstrom Crystal Structure of Putative Oxidoreductase (ycdW) from Salmonella typhimurium | | Descriptor: | Glyoxylate/hydroxypyruvate reductase A | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Wang, Y, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-11-24 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Crystal Structure of Putative Oxidoreductase (ycdW) from Salmonella typhimurium.
TO BE PUBLISHED
|
|
5W1J
 
 | |
3PR2
 
 | | Tryptophan synthase indoline quinonoid structure with F9 inhibitor in alpha site | | Descriptor: | (Z)-N-[(1E)-1-carboxy-2-(2,3-dihydro-1H-indol-1-yl)ethylidene]{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methanaminium, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, CESIUM ION, ... | | Authors: | Lai, J, Niks, D, Wang, Y, Domratcheva, T, Barends, T.R.M, Schwarz, F, Olsen, R.A, Elliott, D.W, Fatmi, M.Q, Chang, C.A, Schlichting, I, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2010-11-29 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray and NMR Crystallography in an Enzyme Active Site: The Indoline Quinonoid Intermediate in Tryptophan Synthase.
J.Am.Chem.Soc., 133, 2011
|
|
5W1L
 
 | |
3OU1
 
 | | MDR769 HIV-1 protease complexed with RH/IN hepta-peptide | | Descriptor: | MDR HIV-1 protease, RH/IN substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
8HNS
 
 | | Crystal structure of an anti-CRISPR protein AcrIIC4 in apo form | | Descriptor: | GLYCEROL, anti-CRISPR protein AcrIIC4 | | Authors: | Sun, W, Cheng, Z, Yang, J, Wang, Y. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HNV
 
 | | CryoEM structure of HpaCas9-sgRNA-dsDNA in the presence of AcrIIC4 | | Descriptor: | CRISPR-associated endonuclease Cas9, anti-CRISPR protein AcrIIC4, non-target strand, ... | | Authors: | Sun, W, Cheng, Z, Wang, J, Yang, X, Wang, Y. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HNW
 
 | | Crystal structure of HpaCas9-sgRNA surveillance complex bound to double-stranded DNA | | Descriptor: | CRISPR-associated endonuclease Cas9, Non-target strand, Target strand, ... | | Authors: | Sun, W, Cheng, Z, Wang, Y. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HNT
 
 | |
3OE7
 
 | | Structure of four mutant forms of yeast f1 ATPase: gamma-I270T | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
J.Biol.Chem., 285, 2010
|
|
3OAI
 
 | | Crystal structure of the extra-cellular domain of human myelin protein zero | | Descriptor: | Maltose-binding periplasmic protein, Myelin protein P0, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Sohi, J, Kamholz, J, Kovari, L.C. | | Deposit date: | 2010-08-05 | | Release date: | 2011-12-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the extracellular domain of human myelin protein zero.
Proteins, 80, 2012
|
|
5W7H
 
 | | Supercharged arPTE variant R5 | | Descriptor: | Phosphotriesterase, ZINC ION | | Authors: | Campbell, E, Grant, J, Wang, Y, Sandhu, M, Williams, R.J, Nisbet, D.R, Perriman, A, Lupton, D, Jackson, C.J. | | Deposit date: | 2017-06-19 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Hydrogel-Immobilized Supercharged Proteins
Adv Biosyst, 2018
|
|
2HVX
 
 | | Discovery of Potent, Orally Active, Nonpeptide Inhibitors of Human Mast Cell Chymase by Using Structure-Based Drug Design | | Descriptor: | Chymase, [(1S)-1-(5-CHLORO-1-BENZOTHIEN-3-YL)-2-(2-NAPHTHYLAMINO)-2-OXOETHYL]PHOSPHONIC ACID | | Authors: | Greco, M.N, Hawkins, M.J, Powell, E.T, Almond, H.R, de Garavilla, L, Wang, Y, Minor, L.A, Wells, G.I, Di Cera, E, Cantwell, A.M, Savvides, S.N, Damiano, B.P, Maryanoff, B.E. | | Deposit date: | 2006-07-31 | | Release date: | 2007-06-12 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of potent, selective, orally active, nonpeptide inhibitors of human mast cell chymase.
J.Med.Chem., 50, 2007
|
|
3OEE
 
 | | Structure of four mutant forms of yeast F1 ATPase: alpha-F405S | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of four mutant forms of yeast F1 ATPase: alpha-F405S
To be Published
|
|
3OFN
 
 | | Structure of four mutant forms of yeast F1 ATPase: alpha-N67I | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-15 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
J.Biol.Chem., 285, 2010
|
|
3QG9
 
 | | crystal structure of FBF-2/gld-1 FBEa A7U mutant complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-R(*UP*GP*UP*GP*CP*CP*UP*UP*A)-3', Fem-3 mRNA-binding factor 2 | | Authors: | Koh, Y.Y, Wang, Y, Qiu, C, Opperman, L, Gross, L, Hall, T.M.T, Wickens, M. | | Deposit date: | 2011-01-24 | | Release date: | 2011-03-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stacking interactions in PUF-RNA complexes.
Rna, 17, 2011
|
|
6KZC
 
 | | crystal structure of TRKc in complex with 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methyl-N-(3-((4- methylpiperazin-1-yl)methyl)-5- (trifluoromethyl)phenyl)benzamide | | Descriptor: | 3-(2-imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methyl-~{N}-[3-(4-methylpiperazin-1-yl)-5-(trifluoromethyl)phenyl]benzamide, NT-3 growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y. | | Deposit date: | 2019-09-23 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis and biological evaluation of 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methylbenzamides as potent and selective pan-tropomyosin receptor kinase (TRK) inhibitors.
Eur.J.Med.Chem., 179, 2019
|
|
4E6K
 
 | | 2.0 A resolution structure of Pseudomonas aeruginosa bacterioferritin (BfrB) in complex with bacterioferritin associated ferredoxin (Bfd) | | Descriptor: | Bacterioferritin, FE2/S2 (INORGANIC) CLUSTER, PHOSPHATE ION, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Wang, Y, Kumar, R, Ruvinsky, A, Vasker, I, Rivera, M. | | Deposit date: | 2012-03-15 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of the BfrB-Bfd Complex Reveals Protein-Protein Interactions Enabling Iron Release from Bacterioferritin.
J.Am.Chem.Soc., 134, 2012
|
|
8JIN
 
 | |
