7QV7
 
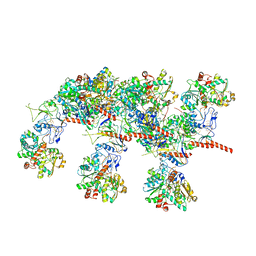 | | Cryo-EM structure of Hydrogen-dependent CO2 reductase. | | Descriptor: | Hydrogen dependent carbon dioxide reductase subunit FdhF, Hydrogen dependent carbon dioxide reductase subunit HycB3, Hydrogen dependent carbon dioxide reductase subunit HycB4, ... | | Authors: | Dietrich, H.M, Righetto, R.D, Kumar, A, Wietrzynski, W, Schuller, S.K, Trischler, R, Wagner, J, Schwarz, F.M, Engel, B.D, Mueller, V, Schuller, J.M. | | Deposit date: | 2022-01-19 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Membrane-anchored HDCR nanowires drive hydrogen-powered CO 2 fixation.
Nature, 607, 2022
|
|
3PR2
 
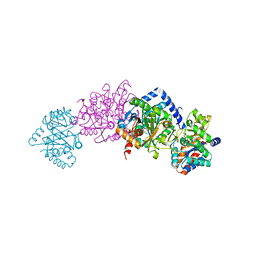 | | Tryptophan synthase indoline quinonoid structure with F9 inhibitor in alpha site | | Descriptor: | (Z)-N-[(1E)-1-carboxy-2-(2,3-dihydro-1H-indol-1-yl)ethylidene]{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methanaminium, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, CESIUM ION, ... | | Authors: | Lai, J, Niks, D, Wang, Y, Domratcheva, T, Barends, T.R.M, Schwarz, F, Olsen, R.A, Elliott, D.W, Fatmi, M.Q, Chang, C.A, Schlichting, I, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2010-11-29 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray and NMR Crystallography in an Enzyme Active Site: The Indoline Quinonoid Intermediate in Tryptophan Synthase.
J.Am.Chem.Soc., 133, 2011
|
|
1XB8
 
 | | Zn substituted form of D62C/K74C double mutant of Pseudomonas Aeruginosa Azurin | | Descriptor: | Azurin, ZINC ION | | Authors: | Tigerstrom, A, Schwarz, F, Karlsson, G, Okvist, M, Alvarez-Rua, C, Maeder, D, Robb, F.T, Sjolin, L. | | Deposit date: | 2004-08-30 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of a novel disulfide bond and engineered electrostatic interactions on the thermostability of azurin
Biochemistry, 43, 2004
|
|
1XB6
 
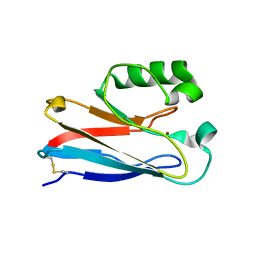 | | The K24R mutant of Pseudomonas Aeruginosa Azurin | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | Tigerstrom, A, Schwarz, F, Karlsson, G, Okvist, M, Alvarez-Rua, C, Maeder, D, Robb, F.T, Sjolin, L. | | Deposit date: | 2004-08-30 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Effects of a novel disulfide bond and engineered electrostatic interactions on the thermostability of azurin
Biochemistry, 43, 2004
|
|
1GZ9
 
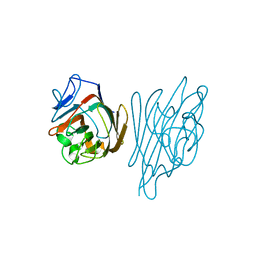 | | High-Resolution Crystal Structure of Erythrina cristagalli Lectin in Complex with 2'-alpha-L-Fucosyllactose | | Descriptor: | CALCIUM ION, ERYTHRINA CRISTA-GALLI LECTIN, MANGANESE (II) ION, ... | | Authors: | Svensson, C, Teneberg, S, Nilsson, C.L, Kjellberg, A, Schwarz, F.P, Sharon, N, Krengel, U. | | Deposit date: | 2002-05-17 | | Release date: | 2002-06-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-Resolution Crystal Structures of Erythrina Cristagalli Lectin in Complex with Lactose and 2'-Alpha-L-Fucosyllactose and Correlation with Thermodynamic Binding Data
J.Mol.Biol., 321, 2002
|
|
1XB3
 
 | | The D62C/K74C double mutant of Pseudomonas Aeruginosa Azurin | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | Tigerstrom, A, Schwarz, F, Karlsson, G, Okvist, M, Alvarez-Rua, C, Maeder, D, Robb, F.T, Sjolin, L. | | Deposit date: | 2004-08-27 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Effects of a novel disulfide bond and engineered electrostatic interactions on the thermostability of azurin
Biochemistry, 43, 2004
|
|
2OUT
 
 | | Solution Structure of HI1506, a Novel Two Domain Protein from Haemophilus influenzae | | Descriptor: | Mu-like prophage FluMu protein gp35, Protein HI1507 in Mu-like prophage FluMu region | | Authors: | Sari, N, He, Y, Doseeva, V, Surabian, K, Schwarz, F, Herzberg, O, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2007-02-12 | | Release date: | 2007-05-01 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HI1506, a novel two-domain protein from Haemophilus influenzae.
Protein Sci., 16, 2007
|
|
