6JIX
 
 | | The cyrstal structure of taurine:2-oxoglutarate aminotransferase from Bifidobacterium kashiwanohense, in complex with PLP and glutamate | | Descriptor: | GLUTAMIC ACID, PYRIDOXAL-5'-PHOSPHATE, taurine:2-oxoglutarate aminotransferase | | Authors: | Li, M, Lin, L, Zhang, Y, Yuchi, Z. | | Deposit date: | 2019-02-23 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.647 Å) | | Cite: | Biochemical and structural investigation of taurine:2-oxoglutarate aminotransferase fromBifidobacterium kashiwanohense.
Biochem.J., 476, 2019
|
|
5WPW
 
 | | Crystal structure of coconut allergen cocosin | | Descriptor: | 11S globulin isoform 1 | | Authors: | Jin, T, Zhang, Y. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Crystal Structure of Cocosin, A Potential Food Allergen from Coconut (Cocos nucifera)
J. Agric. Food Chem., 65, 2017
|
|
8YC0
 
 | | T cell receptor V delta2 V gamma9 in GDN | | Descriptor: | CHOLESTEROL, T cell receptor delta variable 2,T cell receptor delta constant, T cell receptor gamma variable 9,T cell receptor gamma constant 1, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2024-02-17 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
7XZO
 
 | | Formate-tetrahydrofolate ligase in complex with ATP | | Descriptor: | (R,R)-2,3-BUTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Formate--tetrahydrofolate ligase, ... | | Authors: | Fang, C.L, Zhang, Y. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of FtfL as a novel target of berberine in intestinal bacteria.
Bmc Biol., 21, 2023
|
|
7XZN
 
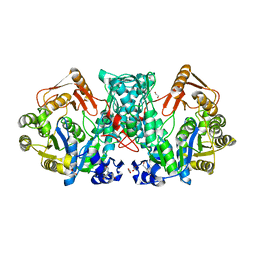 | |
7XZP
 
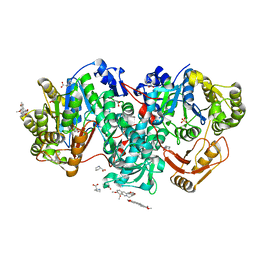 | |
7KOJ
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder494 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]-5-{[(prop-2-en-1-yl)carbamoyl]amino}benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder494
to be published
|
|
7KOK
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder496 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder496
to be published
|
|
7YL2
 
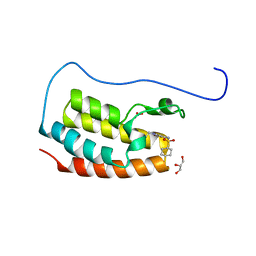 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004 | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, N-(1-ethyl-2-oxidanylidene-3H-indol-5-yl)cyclohexanesulfonamide, ... | | Authors: | Huang, Y, Wei, A, Dong, R, Xu, H, Zhang, C, Chen, Z, Li, J, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-07-25 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004
To Be Published
|
|
7JN2
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder441 inhibitor | | Descriptor: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-03 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder441
to be published
|
|
6IEX
 
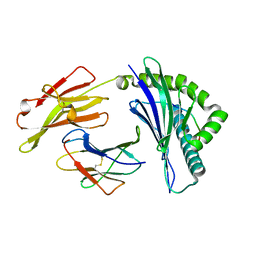 | | Crystal structure of HLA-B*4001 in complex with SARS-CoV derived peptide N216-225 GETALALLLL | | Descriptor: | Beta-2-microglobulin, GLY-GLU-THR-ALA-LEU-ALA-LEU-LEU-LEU-LEU, MHC class I antigen | | Authors: | Ji, W, Niu, L, Peng, W, Zhang, Y, Shi, Y, Qi, J, Gao, G.F, Liu, W.J. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.314 Å) | | Cite: | Salt bridge-forming residues positioned over viral peptides presented by MHC class I impacts T-cell recognition in a binding-dependent manner.
Mol.Immunol., 112, 2019
|
|
7JJO
 
 | | Structural Basis of the Activation of Heterotrimeric Gs-protein by Isoproterenol-bound Beta1-Adrenergic Receptor | | Descriptor: | Beta1-Adrenergic Receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Su, M, Zhu, L, Zhang, Y, Paknejad, N, Dey, R, Huang, J, Lee, M.Y, Williams, D, Jordan, K.D, Eng, E.T, Ernst, O.P, Meyerson, J.R, Hite, R.K, Walz, T, Liu, W, Huang, X.Y. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural Basis of the Activation of Heterotrimeric Gs-Protein by Isoproterenol-Bound beta 1 -Adrenergic Receptor.
Mol.Cell, 80, 2020
|
|
5ZIT
 
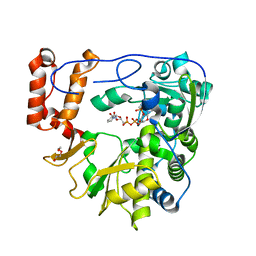 | | Crystal structure of human Enterovirus D68 RdRp in complex with NADPH | | Descriptor: | DI(HYDROXYETHYL)ETHER, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, RdRp | | Authors: | Wang, M.L, Li, L, Chen, Y.P, Jiang, H, Zhang, Y, Su, D. | | Deposit date: | 2018-03-17 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 2020
|
|
7KRX
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441 inhibitor | | Descriptor: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441
to be published
|
|
7KOL
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496
to be published
|
|
2FF0
 
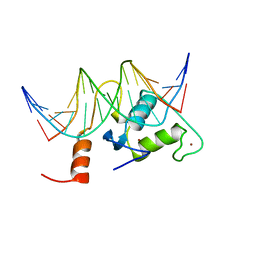 | | Solution Structure of Steroidogenic Factor 1 DNA Binding Domain Bound to its Target Sequence in the Inhibin alpha-subunit Promoter | | Descriptor: | CTGTGGCCCTGAGCC, GGCTCAGGGCCACAG, Steroidogenic factor 1, ... | | Authors: | Little, T.H, Zhang, Y, Matulis, C.K, Weck, J, Zhang, Z, Ramachandran, A, Mayo, K.E, Radhakrishnan, I. | | Deposit date: | 2005-12-17 | | Release date: | 2006-04-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific deoxyribonucleic Acid (DNA) recognition by steroidogenic factor 1: a helix at the carboxy terminus of the DNA binding domain is necessary for complex stability.
Mol.Endocrinol., 20, 2006
|
|
6IVK
 
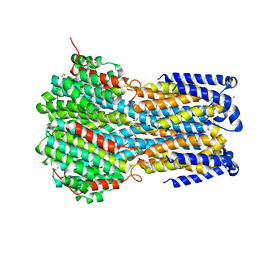 | | Crystal structure of a membrane protein G175A | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
7XW9
 
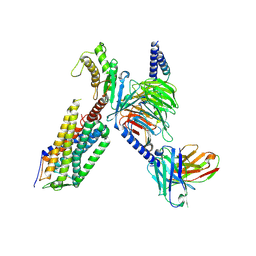 | | Cryo-EM structure of the TRH-bound human TRHR-Gq complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Ji, S, Dong, Y, Chen, L, Zang, S, Shen, D, Guo, J, Qin, J, Zhang, H, Wang, W, Shen, Q, Mao, C, Zhang, Y. | | Deposit date: | 2022-05-26 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis for the activation of thyrotropin-releasing hormone receptor.
Cell Discov, 8, 2022
|
|
6IVN
 
 | | Crystal structure of a membrane protein G264A | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, Ibestrophin, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IVL
 
 | | Crystal structure of a membrane protein L259A | | Descriptor: | ACETIC ACID, CHLORIDE ION, Ibestrophin, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IVJ
 
 | | Crystal structure of a membrane protein G18A | | Descriptor: | ACETIC ACID, CHLORIDE ION, Ibestrophin, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6J1M
 
 | |
6J1N
 
 | | Anisodus acutangulus type III polyketide sythase AaPKS2 in complex with 4-carboxy-3-oxobutanoyl-CoA | | Descriptor: | (3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-10,14,19,21-tetraoxo-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatricosan-23-oic acid 3,5-dioxide (non-preferred name), A. acutangulus PKS2 | | Authors: | Fang, C.L, Zhang, Y. | | Deposit date: | 2018-12-28 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Tropane alkaloids biosynthesis involves an unusual type III polyketide synthase and non-enzymatic condensation.
Nat Commun, 10, 2019
|
|
6JBQ
 
 | |
6IVP
 
 | | Crystal structure of a membrane protein P262A | | Descriptor: | CHLORIDE ION, ZINC ION, bestrophin | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
