5EPY
 
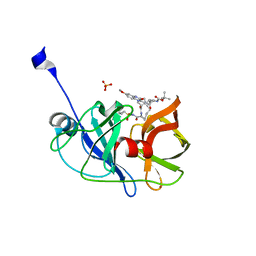 | | Crystal structure of HCV NS3/4A protease A156T variant in complex with 5172-mcP1P3 (MK-5172 P1-P3 macrocyclic analogue) | | Descriptor: | 2-Methyl-2-propanyl {(2R,6S,12Z,13aS,14aR,16aS)-14a-[(cyclopropylsulfonyl)carbamoyl]-2-[(3-ethyl-7-methoxy-2-quinoxalinyl)oxy]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclop ropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, SULFATE ION, ... | | Authors: | Soumana, D.I, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Aydin, C, Schiffer, C.A. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172.
Acs Chem.Biol., 11, 2016
|
|
6RHG
 
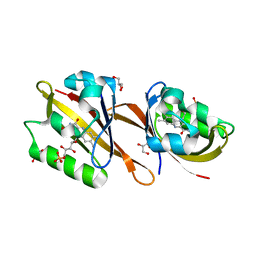 | | Structure of Chloroflexus aggregans Cagg_3753 LOV domain | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase | | Authors: | Nazarenko, V.V, Remeeva, A, Yudenko, A, Kovalev, K, Gordeliy, V, Gushchin, I. | | Deposit date: | 2019-04-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | A thermostable flavin-based fluorescent protein from Chloroflexus aggregans: a framework for ultra-high resolution structural studies.
Photochem. Photobiol. Sci., 18, 2019
|
|
5EKT
 
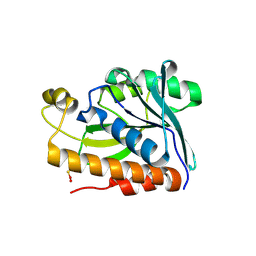 | |
7YX5
 
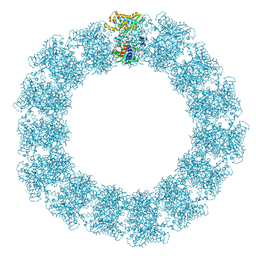 | | Structure of the Mimivirus genomic fibre in its relaxed 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
5ZUR
 
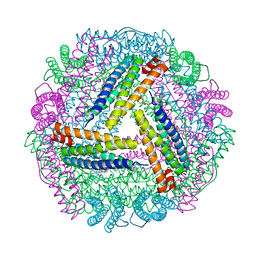 | | Achromobacter Dh1f Bacterioferritin | | Descriptor: | BARIUM ION, Bacterioferritin, CHLORIDE ION, ... | | Authors: | Dwivedy, A, Jha, B, Singh, K.H, Ahmed, M, Ashraf, A, Kumar, D, Biswal, B.K. | | Deposit date: | 2018-05-08 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Serendipitous crystallization and structure determination of bacterioferritin from Achromobacter.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7YX4
 
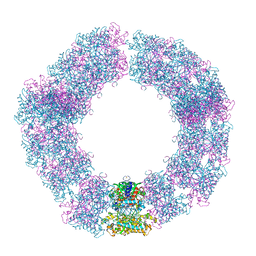 | | Structure of the Mimivirus genomic fibre in its compact 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
5HTB
 
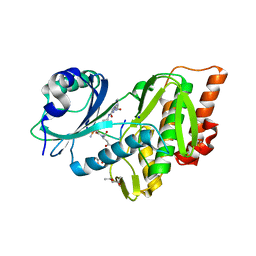 | | Crystal structure of haspin (GSG2) in complex with bisubstrate inhibitor ARC-3353 | | Descriptor: | (3R)-4-amino-3-{[6-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]carbonyl}amino)hexanoyl]amino}-4-oxobutanoic acid (non-preferred name), (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Chaikuad, A, Heroven, C, Lavogina, D, Kestav, K, Uri, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-01-26 | | Release date: | 2016-05-11 | | Last modified: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-crystal structures of the protein kinase haspin with bisubstrate inhibitors.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4WUQ
 
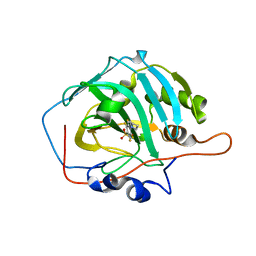 | | Crystal structure of human carbonic anhydrase isozyme I with 2,3,5,6-Tetrafluoro-4-piperidin-1-ylbenzenesulfonamide | | Descriptor: | 2,3,5,6-tetrafluoro-4-(piperidin-1-yl)benzenesulfonamide, Carbonic anhydrase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2014-11-03 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Intrinsic thermodynamics of 4-substituted-2,3,5,6-tetrafluorobenzenesulfonamide binding to carbonic anhydrases by isothermal titration calorimetry.
Biophys.Chem., 205, 2015
|
|
7YX3
 
 | | Structure of the Mimivirus genomic fibre in its compact 6-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30-nm diameter helical protein shield.
Elife, 11, 2022
|
|
8HHZ
 
 | |
5HYK
 
 | | Crystal structure of the complex PPARalpha/AL26-29 | | Descriptor: | 2-methyl-2-[4-(naphthalen-1-yl)phenoxy]propanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Loiodice, F, Laghezza, A, Lavecchia, A. | | Deposit date: | 2016-02-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for PPAR partial or full activation revealed by a novel ligand binding mode.
Sci Rep, 6, 2016
|
|
7OLW
 
 | |
8HHY
 
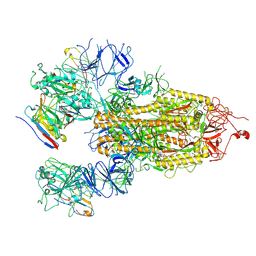 | | SARS-CoV-2 Delta Spike in complex with IS-9A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IS-9A Fab heavy chain, ... | | Authors: | Mohapatra, A, Wu, Y.-M. | | Deposit date: | 2022-11-17 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
3HU7
 
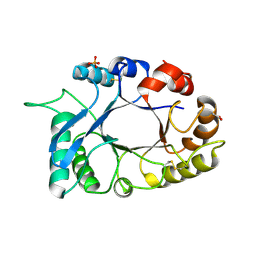 | | Structural characterization and binding studies of a plant pathogenesis related protein heamanthin from haemanthus multiflorus reveal its dual inhibitory effects against xylanase and alpha-amylase | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION | | Authors: | Kumar, S, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-06-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure determination and inhibition studies of a novel xylanase and alpha-amylase inhibitor protein (XAIP) from Scadoxus multiflorus.
Febs J., 277, 2010
|
|
8HPK
 
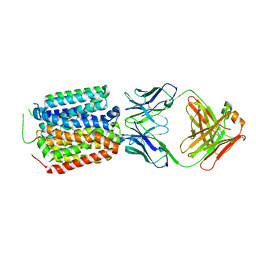 | | Crystal structure of the bacterial oxalate transporter OxlT in an oxalate-bound occluded form | | Descriptor: | Fab fragment Heavy chein, Fab fragment Light chain, OXALATE ION, ... | | Authors: | Shimamura, T, Hirai, T, Yamashita, A. | | Deposit date: | 2022-12-12 | | Release date: | 2023-02-15 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and mechanism of oxalate transporter OxlT in an oxalate-degrading bacterium in the gut microbiota.
Nat Commun, 14, 2023
|
|
8HPJ
 
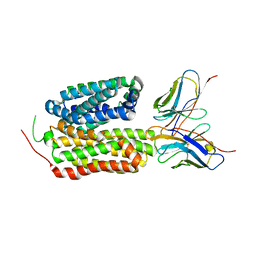 | |
5LBQ
 
 | | LSD1-CoREST1 in complex with quinazoline-derivative reversible inhibitor | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, N2-(3-(dimethylamino)propyl)-6,7-dimethoxy-N4,N4-dimethylquinazoline-2,4-diamine, ... | | Authors: | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | Deposit date: | 2016-06-16 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
4X93
 
 | | Crystal structure of Lysosomal Phospholipase A2 crystallized in the presence of methyl arachidonyl fluorophosphonate (tetragonal form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Glukhova, A, Tesmer, J.J.G. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of lysosomal phospholipase A2 and lecithin:cholesterol acyltransferase.
Nat Commun, 6, 2015
|
|
7OLU
 
 | |
6RPB
 
 | | Crystal structure of the T-cell receptor NYE_S1 bound to HLA A2*01-SLLMWITQV | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Coles, C.H, Mulvaney, R, Malla, S, Lloyd, A, Smith, K, Chester, F, Knox, A, Stacey, A.R, Dukes, J, Baston, E, Griffin, S, Vuidepot, A, Jakobsen, B.K, Harper, S. | | Deposit date: | 2019-05-14 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | TCRs with Distinct Specificity Profiles Use Different Binding Modes to Engage an Identical Peptide-HLA Complex.
J Immunol., 204, 2020
|
|
6RP9
 
 | | Crystal structure of the T-cell receptor NYE_S3 bound to HLA A2*01-SLLMWITQV | | Descriptor: | Beta-2-microglobulin, Cancer/testis antigen 1, HLA class I histocompatibility antigen, ... | | Authors: | Coles, C.H, Mulvaney, R, Malla, S, Lloyd, A, Smith, K, Chester, F, Knox, A, Stacey, A.R, Dukes, J, Baston, E, Griffin, S, Vuidepot, A, Jakobsen, B.K, Harper, S. | | Deposit date: | 2019-05-14 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | TCRs with Distinct Specificity Profiles Use Different Binding Modes to Engage an Identical Peptide-HLA Complex.
J Immunol., 204, 2020
|
|
8CAV
 
 | | Discovery of the lanthipeptide Curvocidin and structural insights into its trifunctional synthetase CuvL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CuvA, MAGNESIUM ION, ... | | Authors: | Sigurdsson, A, Martins, B.M, Duettmann, S.A, Jasyk, M, Dimos-Roehl, B, Schoepf, F, Gemannter, M, Knittel, C.H, Schnegotyzki, R, Schmid, B, Kosol, S, Gonzalez-Viegas, M, Seidel, M, Huegelland, M, Leimkuehler, S, Dobbek, H, Mainz, A, Suessmuth, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Discovery of the Lanthipeptide Curvocidin and Structural Insights into its Trifunctional Synthetase CuvL.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7P4E
 
 | | Crystal structure of PPARgamma in complex with compound FL217 | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma, SULFATE ION, ... | | Authors: | Ni, X, Lillich, F, Proschak, E, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of Dual Partial Peroxisome Proliferator-Activated Receptor gamma Agonists/Soluble Epoxide Hydrolase Inhibitors.
J.Med.Chem., 64, 2021
|
|
5LE2
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DDD_D12_15_D12_15_D12 | | Descriptor: | ACETATE ION, DDD_D12_15_D12_15_D12, THIOCYANATE ION | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LEC
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DDD_D12_12_D12_12_D12 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DDD_D12_12_D12_12_D12 | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
