5O9F
 
 | | Crystal structure of R. ruber ADH-A, mutant Y294F, W295A, Y54F, F43S, H39Y | | Descriptor: | (2~{S})-2-methylpentanedioic acid, Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Relaxation of nonproductive binding and increased rate of coenzyme release in an alcohol dehydrogenase increases turnover with a nonpreferred alcohol enantiomer.
FEBS J., 284, 2017
|
|
5MUB
 
 | | ACC1 Fab fragment in complex with citrullinated C1 epitope of CII (CG05) | | Descriptor: | ACC1 Fab fragment heavy chain, ACC1 Fab fragment light chain, triple-helical peptide containing the citrullinated C1 epitope of collagen type II,Collagen alpha-1(II) chain,triple-helical peptide containing the citrullinated C1 epitope of collagen type II | | Authors: | Dobritzsch, D, Holmdahl, R, Ge, C. | | Deposit date: | 2017-01-13 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Anti-citrullinated protein antibodies cause arthritis by cross-reactivity to joint cartilage.
JCI Insight, 2, 2017
|
|
5MU2
 
 | | ACC1 Fab fragment in complex with CII583-591 (CG10) | | Descriptor: | ACC1 Fab fragment heavy chain, ACC1 Fab fragment light chain, synthetic peptide containing the CII583-591 epitope of collagen type II | | Authors: | Dobritzsch, D, Holmdahl, R, Ge, C. | | Deposit date: | 2017-01-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Anti-citrullinated protein antibodies cause arthritis by cross-reactivity to joint cartilage.
JCI Insight, 2, 2017
|
|
5MU0
 
 | | ACC1 Fab fragment in complex with citrullinated C1 epitope of CII (IA03) | | Descriptor: | IA03 peptide containing the citrullinated C1 epitope of collagen type II,Collagen alpha-1(II) chain,IA03 peptide containing the citrullinated C1 epitope of collagen type II, heavy chain of ACC1 antibody Fab fragment, light chain of ACC1 antibody Fab fragment | | Authors: | Dobritzsch, D, Holmdahl, R, Ge, C. | | Deposit date: | 2017-01-11 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Anti-citrullinated protein antibodies cause arthritis by cross-reactivity to joint cartilage.
JCI Insight, 2, 2017
|
|
5MV3
 
 | | ACC1 Fab fragment in complex with CII583-591 (CG10) | | Descriptor: | heavy chain of ACC1 Fab fragment, light chain of ACC1 Fab fragment, synthetic peptide containing the CII583-591 epitope of collagen type II,Collagen alpha-1(II) chain,synthetic peptide containing the CII583-591 epitope of collagen type II, | | Authors: | Dobritzsch, D, Holmdahl, R, Ge, C. | | Deposit date: | 2017-01-15 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Anti-citrullinated protein antibodies cause arthritis by cross-reactivity to joint cartilage.
JCI Insight, 2, 2017
|
|
5MV4
 
 | | ACC1 Fab fragment in complex with citrullinated CII616-639 epitope of collagen type II (ptm23) | | Descriptor: | ACC1 antibody Fab fragment, heavy chain, light chain, ... | | Authors: | Dobritzsch, D, Holmdahl, R, Ge, C. | | Deposit date: | 2017-01-15 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Anti-citrullinated protein antibodies cause arthritis by cross-reactivity to joint cartilage.
JCI Insight, 2, 2017
|
|
5OD8
 
 | | Crystal structure of the RA-associated mAb B2 (Fab fragment) | | Descriptor: | B2 Fab fragment - Light chain, B2 Fab fragment - heavy chain | | Authors: | Dobritzsch, D, Ge, C, Holmdahl, R, Amara, K, Malmstrom, V. | | Deposit date: | 2017-07-05 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies.
Arthritis Rheumatol, 71, 2019
|
|
5OD0
 
 | | Crystal structure of ACPA E4 | | Descriptor: | Fab fragment of ACPA E4 - Light chain, Fab fragment of ACPA E4 - heavy chain, GLYCEROL | | Authors: | Dobritzsch, D, Ge, C, Holmdahl, R. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies.
Arthritis Rheumatol, 71, 2019
|
|
5OCY
 
 | |
5OCK
 
 | | Crystal structure of ACPA E4 in complex with CEP1 | | Descriptor: | CEP1 peptide (from enolase), Human ACPA E4 Fab fragment - Heavy chain, Human ACPA E4 Fab fragment - Light chain | | Authors: | Dobritzsch, D, Ge, C, Holmdahl, R. | | Deposit date: | 2017-07-03 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies.
Arthritis Rheumatol, 71, 2019
|
|
5ODB
 
 | | Crystal structure of the RA-associated mAb D10 (chimeric Fab fragment) | | Descriptor: | D10 Fab fragment - heavy chain, D10 Fab fragment - light chain, GLYCEROL, ... | | Authors: | Dobritzsch, D, Ge, C, Holmdahl, R, Amara, K, Malmstrom, V. | | Deposit date: | 2017-07-05 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies.
Arthritis Rheumatol, 71, 2019
|
|
5OCX
 
 | | Crystal structure of ACPA E4 in complex with CII-C-13-CIT | | Descriptor: | 1,2-ETHANEDIOL, CII-C-13-CIT, Fab fragment anti-citrullinated protein antibody E4 - light chain, ... | | Authors: | Dobritzsch, D, Ge, C, Holmdahl, R. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies.
Arthritis Rheumatol, 71, 2019
|
|
8BBR
 
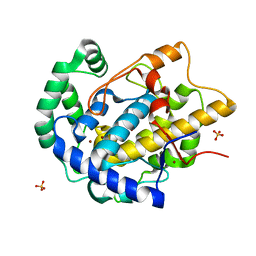 | | Determination of the structure of active tyrosinase from bacterium Verrucomicrobium spinosum | | Descriptor: | COPPER (II) ION, Core tyrosinase, SULFATE ION | | Authors: | Fekry, M, Dave, K, Badgujar, D, Aurelius, O, Hamnevik, E, Dobritzsch, D, Danielson, H. | | Deposit date: | 2022-10-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Crystal Structure of Tyrosinase from Verrucomicrobium spinosum Reveals It to Be an Atypical Bacterial Tyrosinase.
Biomolecules, 13, 2023
|
|
2R6S
 
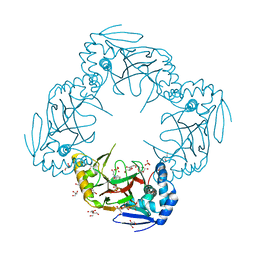 | | Crystal structure of Gab protein | | Descriptor: | BICINE, FE (II) ION, GLYCEROL, ... | | Authors: | Lohkamp, B, Dobritzsch, D. | | Deposit date: | 2007-09-06 | | Release date: | 2008-06-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A mixture of fortunes: the curious determination of the structure of Escherichia coli BL21 Gab protein.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6YUH
 
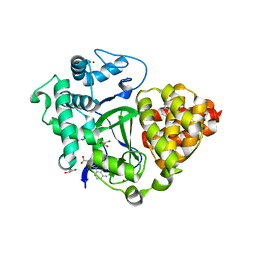 | | Crystal structure of SMYD3 with diperodon R enantiomer bound to allosteric site | | Descriptor: | Diperodon, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Cederfelt, D, Talibov, V.O, Dobritzsch, D, Danielson, U.H. | | Deposit date: | 2020-04-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of an Allosteric Ligand Binding Site in SMYD3 Lysine Methyltransferase.
Chembiochem, 22, 2021
|
|
7BJ1
 
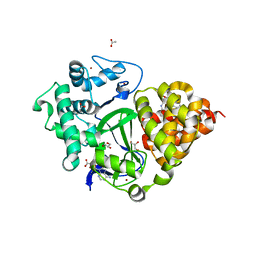 | | Crystal structure of SMYD3 with diperodon S enantiomer bound to allosteric site | | Descriptor: | ACETATE ION, Diperodon (S-enantiomer), GLYCEROL, ... | | Authors: | Talibov, V.O, Cederfelt, D, Dobritzsch, D, Danielson, U.H. | | Deposit date: | 2021-01-13 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Discovery of an Allosteric Ligand Binding Site in SMYD3 Lysine Methyltransferase
Chembiochem, 22, 2021
|
|
8PT4
 
 | | beta-Ureidopropionase tetramer | | Descriptor: | Beta-ureidopropionase | | Authors: | Cederfelt, D, Dobritzsch, D. | | Deposit date: | 2023-07-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | The Allosteric Regulation of Beta-Ureidopropionase Depends on Fine-Tuned Stability of Active-Site Loops and Subunit Interfaces.
Biomolecules, 13, 2023
|
|
8BBQ
 
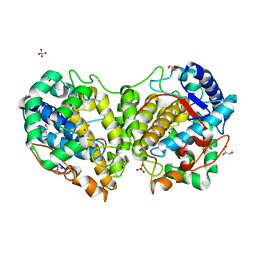 | | Determination of the structure of active tyrosinase from bacterium Verrucomicrobium spinosum | | Descriptor: | COPPER (II) ION, Core tyrosinase, GLYCEROL, ... | | Authors: | Fekry, M, Dave, K, Badgujar, D, Aurelius, O, Hamnevik, E, Dobritzsch, D, Danielson, H. | | Deposit date: | 2022-10-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The Crystal Structure of Tyrosinase from Verrucomicrobium spinosum Reveals It to Be an Atypical Bacterial Tyrosinase.
Biomolecules, 13, 2023
|
|
1QPB
 
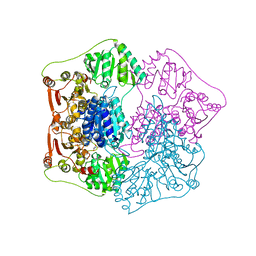 | | PYRUVATE DECARBOYXLASE FROM YEAST (FORM B) COMPLEXED WITH PYRUVAMIDE | | Descriptor: | MAGNESIUM ION, PYRUVAMIDE, PYRUVATE DECARBOXYLASE (FORM B), ... | | Authors: | Lu, G, Dobritzsch, D, Schneider, G. | | Deposit date: | 1999-11-26 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structural Basis of Substrate Activation in Yeast Pyruvate Decarboxylase a Crystallographic and Kinetic Study
Eur.J.Biochem., 267, 2000
|
|
2FTW
 
 | |
1R43
 
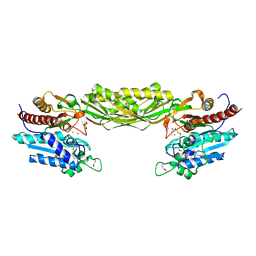 | | Crystal structure of beta-alanine synthase from Saccharomyces kluyveri (selenomethionine substituted protein) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-AMINO ISOBUTYRATE, ... | | Authors: | Lundgren, S, Gojkovic, Z, Piskur, J, Dobritzsch, D. | | Deposit date: | 2003-10-03 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Yeast beta-Alanine Synthase Shares a Structural Scaffold and Origin with Dizinc-dependent Exopeptidases
J.Biol.Chem., 278
|
|
1R3N
 
 | | Crystal structure of beta-alanine synthase from Saccharomyces kluyveri | | Descriptor: | BETA-AMINO ISOBUTYRATE, ZINC ION, beta-alanine synthase | | Authors: | Lundgren, S, Gojkovic, Z, Piskur, J, Dobritzsch, D. | | Deposit date: | 2003-10-02 | | Release date: | 2003-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Yeast beta-Alanine Synthase Shares a Structural Scaffold and Origin with Dizinc-dependent Exopeptidases
J.Biol.Chem., 278, 2003
|
|
2VL1
 
 | | Crystal structure of beta-alanine synthase from Saccharomyces kluyveri in complex with a gly-gly peptide | | Descriptor: | BETA-ALANINE SYNTHASE, GLYCINE, ZINC ION | | Authors: | Andersen, B, Lundgren, S, Dobritzsch, D, Piskur, J. | | Deposit date: | 2008-01-07 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Recruited Protease is Involved in Catabolism of Pyrimidines.
J.Mol.Biol., 379, 2008
|
|
7ZX4
 
 | | Clathrin N-terminal domain in complex with a HURP phospho-peptide | | Descriptor: | CHLORIDE ION, Clathrin heavy chain 1, Disks large-associated protein 5, ... | | Authors: | Kliche, J, Badgujar, D, Dobritzsch, D, Ivarsson, Y. | | Deposit date: | 2022-05-20 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Large-scale phosphomimetic screening identifies phospho-modulated motif-based protein interactions.
Mol.Syst.Biol., 19, 2023
|
|
7NDV
 
 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL001888. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[4-(trifluoromethyl)phenoxy]piperidine, Acetylcholine-binding protein, ... | | Authors: | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | Deposit date: | 2021-02-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of fragments inducing conformational effects in dynamic proteins using a second-harmonic generation biosensor
RSC Advances, 11, 2021
|
|
