3PBQ
 
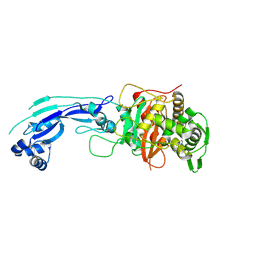 | | Crystal structure of PBP3 complexed with imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2011-08-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PD9
 
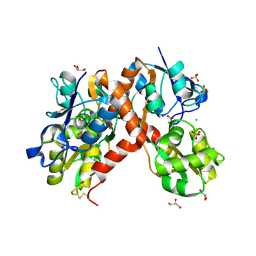 | | X-ray structure of the ligand-binding core of GluA2 in complex with (R)-5-HPCA at 2.1 A resolution | | Descriptor: | (5R)-3-hydroxy-4,5,6,7-tetrahydroisoxazolo[5,4-c]pyridine-5-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biostructural and pharmacological studies of bicyclic analogues of the 3-isoxazolol glutamate receptor agonist ibotenic acid.
J. Med. Chem., 53, 2010
|
|
3PBR
 
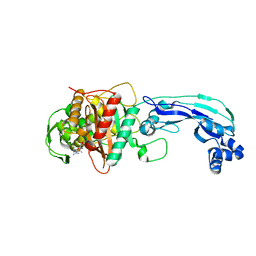 | | Crystal structure of PBP3 complexed with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2012-05-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8QFY
 
 | |
7KLM
 
 | | Human Arginase1 Complexed with Inhibitor Compound 24a | | Descriptor: | 3-[(2~{S},3~{R},4~{R})-4-[[(2~{S})-2-azanyl-3-methyl-butanoyl]amino]-2-carboxy-pyrrolidin-3-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boron, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-Based Discovery of Proline-Derived Arginase Inhibitors with Improved Oral Bioavailability for Immuno-Oncology.
Acs Med.Chem.Lett., 12, 2021
|
|
7KLL
 
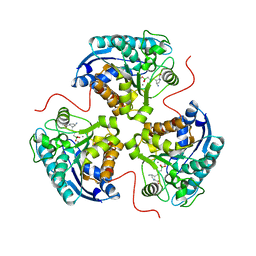 | | Human Arginase1 Complexed with Inhibitor Compound 18 | | Descriptor: | 3-[(2~{S},3~{R},4~{R})-4-azanyl-2-carboxy-pyrrolidin-3-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boron, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-Based Discovery of Proline-Derived Arginase Inhibitors with Improved Oral Bioavailability for Immuno-Oncology.
Acs Med.Chem.Lett., 12, 2021
|
|
1BWH
 
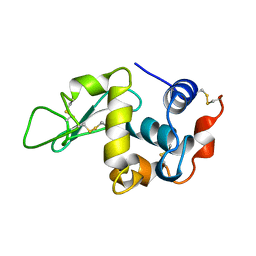 | | THE 1.8 A STRUCTURE OF GROUND CONTROL GROWN TETRAGONAL HEN EGG WHITE LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME) | | Authors: | Dong, J, Boggon, T.J, Chayen, N.E, Raftery, J, Bi, R.C. | | Deposit date: | 1998-09-24 | | Release date: | 1998-09-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bound-solvent structures for microgravity-, ground control-, gel- and microbatch-grown hen egg-white lysozyme crystals at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BRE
 
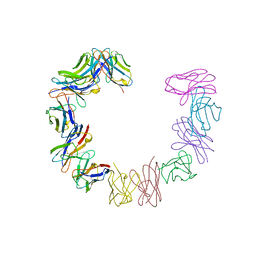 | | IMMUNOGLOBULIN LIGHT CHAIN PROTEIN | | Descriptor: | BENCE-JONES KAPPA I PROTEIN BRE | | Authors: | Schormann, N, Benson, M.D. | | Deposit date: | 1995-07-19 | | Release date: | 1995-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tertiary structure of an amyloid immunoglobulin light chain protein: a proposed model for amyloid fibril formation.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
7JUZ
 
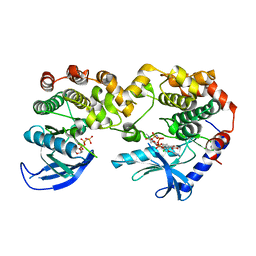 | |
7JRA
 
 | |
7JUQ
 
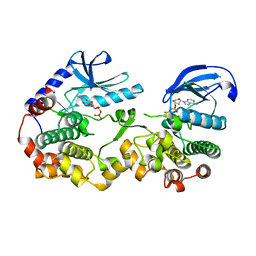 | | Crystal Structure of KSR2:MEK1 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, Kinase suppressor of Ras 2, ... | | Authors: | Khan, Z.M, Dar, A.C. | | Deposit date: | 2020-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural basis for the action of the drug trametinib at KSR-bound MEK.
Nature, 588, 2020
|
|
7JUR
 
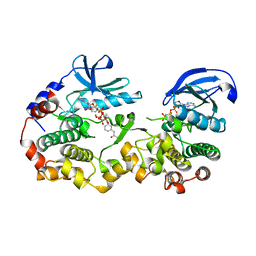 | |
7JUX
 
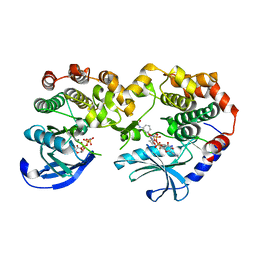 | |
7JUY
 
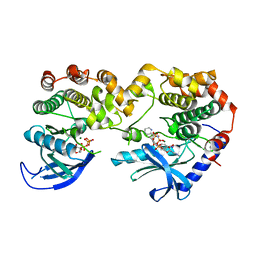 | |
7JZ6
 
 | |
8S9K
 
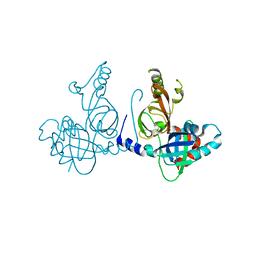 | | Structure of dimeric FAM111A SPD S541A Mutant | | Descriptor: | GLYCEROL, Serine protease FAM111A | | Authors: | Palani, S, Alvey, J.A, Cong, A.T.Q, Schellenberg, M.J, Machida, Y. | | Deposit date: | 2023-03-29 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Dimerization-dependent serine protease activity of FAM111A prevents replication fork stalling at topoisomerase 1 cleavage complexes.
Nat Commun, 15, 2024
|
|
7JUU
 
 | |
7KLK
 
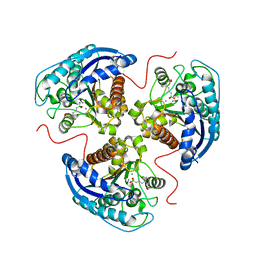 | | Human Arginase1 Complexed with Inhibitor Compound 3a | | Descriptor: | 1,2-ETHANEDIOL, 3-[(2~{S},3~{R})-2-carboxypiperidin-3-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, ... | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure-Based Discovery of Proline-Derived Arginase Inhibitors with Improved Oral Bioavailability for Immuno-Oncology.
Acs Med.Chem.Lett., 12, 2021
|
|
8S9L
 
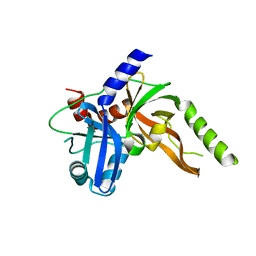 | | Structure of monomeric FAM111A SPD V347D Mutant | | Descriptor: | SULFATE ION, Serine protease FAM111A | | Authors: | Palani, S, Alvey, J.A, Cong, A.T.Q, Schellenberg, M.J, Machida, Y. | | Deposit date: | 2023-03-29 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dimerization-dependent serine protease activity of FAM111A prevents replication fork stalling at topoisomerase 1 cleavage complexes.
Nat Commun, 15, 2024
|
|
7L0L
 
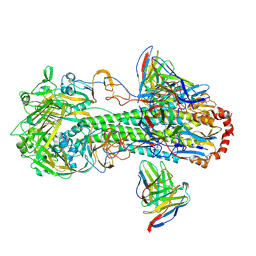 | | Cryo-EM structure of the VRC316 clinical trial, vaccine-elicited, human antibody 316-310-1B11 in complex with an H2 CAN05 HA trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 316-310-1B11 Heavy Chain, 316-310-1B11 Light Chain, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-11-03 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | A single residue in influenza virus H2 hemagglutinin enhances the breadth of the B cell response elicited by H2 vaccination.
Nat Med, 28, 2022
|
|
1E26
 
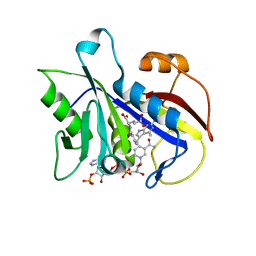 | | Design, Synthesis and X-ray Crystal Structure of a Potent Dual Inhibitor of Thymidylate Synthase and Dihydrofolate Reductase as an Antitumor Agent. | | Descriptor: | DIHYDROFOLATE REDUCTASE, N-[4-[2-(2-AMINO-4-METHYL-7H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYL]GLUTAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gangjee, A, Yu, J, McGuire, J.J, Cody, V, Galitsky, N, Kisliuk, R.L, Queener, S.F. | | Deposit date: | 2000-05-17 | | Release date: | 2001-05-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand-Induced Conformational Changes in the Crystal Structures of Pneumocystis Carinii Dihydrofolate Reductase Complexes with Folate and Nadp+
Biochemistry, 38, 1999
|
|
9CWV
 
 | |
9D6C
 
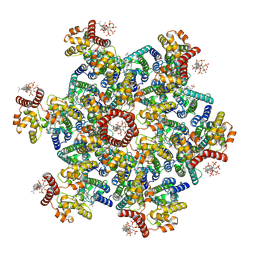 | | Gag CA-SP1 immature lattice bound with Lenacapavir and Bevirimat from enveloped virus like particles | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Wu, C, Meuser, M.E, Xiong, Y. | | Deposit date: | 2024-08-14 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structural insights into inhibitor mechanisms on immature HIV-1 Gag lattice revealed by high-resolution in situ single-particle cryo-EM.
Biorxiv, 2024
|
|
9D6D
 
 | | Gag CA-SP1 immature lattice bound with Lenacapavir from enveloped virus like particles | | Descriptor: | Gag polyprotein, INOSITOL HEXAKISPHOSPHATE, N-[(1S)-1-(3-{4-chloro-3-[(methylsulfonyl)amino]-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl}-6-[3-methyl-3-(methylsulfonyl)but-1-yn-1-yl]pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-5,5-difluoro-3-(trifluoromethyl)-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Wu, C, Meuser, M.E, Xiong, Y. | | Deposit date: | 2024-08-14 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Structural insights into inhibitor mechanisms on immature HIV-1 Gag lattice revealed by high-resolution in situ single-particle cryo-EM.
Biorxiv, 2024
|
|
9D6E
 
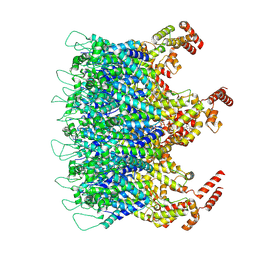 | | Gag CA-SP1 immature lattice bound with Bevirimat from enveloped virus like particles | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Wu, C, Meuser, M.E, Xiong, Y. | | Deposit date: | 2024-08-14 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural insights into inhibitor mechanisms on immature HIV-1 Gag lattice revealed by high-resolution in situ single-particle cryo-EM.
Biorxiv, 2024
|
|
