6PWV
 
 | | Cryo-EM structure of MLL1 core complex bound to the nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
7CFN
 
 | | Cryo-EM structure of the INT-777-bound GPBAR-Gs complex | | Descriptor: | (2S,4R)-4-[(3R,5S,6R,7R,8R,9S,10S,12S,13R,14S,17R)-6-ethyl-10,13-dimethyl-3,7,12-tris(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-pentanoic acid, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
4U7D
 
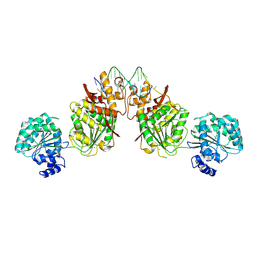 | | Structure of human RECQ-like helicase in complex with an oligonucleotide | | Descriptor: | ATP-dependent DNA helicase Q1, DNA oligonucleotide, ZINC ION | | Authors: | Pike, A.C.W, Zhang, Y, Schnecke, C, Cooper, C.D.O, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-30 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Human RECQ1 helicase-driven DNA unwinding, annealing, and branch migration: Insights from DNA complex structures.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7CX4
 
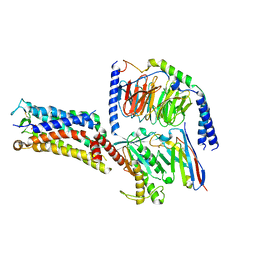 | | Cryo-EM structure of the Evatanepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-~{tert}-butylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7CX3
 
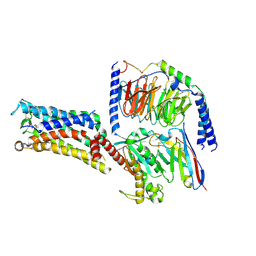 | | Cryo-EM structure of the Taprenepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-pyrazol-1-ylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7CX2
 
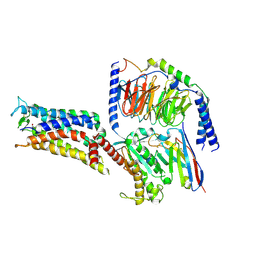 | | Cryo-EM structure of the PGE2-bound EP2-Gs complex | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
6EDR
 
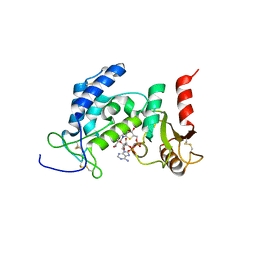 | | Crystal Structure of Human CD38 in Complex with 4'-Thioribose NAD+ | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, [[(2~{R},3~{S},4~{R},5~{R})-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)thiolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Dai, Z, Zhang, X.N, Nasertorabi, F, Cheng, Q, Pei, H, Louie, S.G, Stevens, C.R, Zhang, Y. | | Deposit date: | 2018-08-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Facile chemoenzymatic synthesis of a novel stable mimic of NAD.
Chem Sci, 9, 2018
|
|
1FRP
 
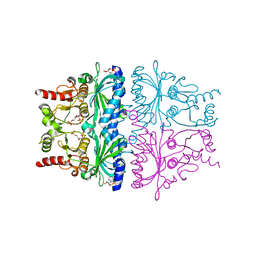 | | CRYSTAL STRUCTURE OF FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH FRUCTOSE-2,6-BISPHOSPHATE, AMP AND ZN2+ AT 2.0 ANGSTROMS RESOLUTION. ASPECTS OF SYNERGISM BETWEEN INHIBITORS | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Xue, Y, Huang, S, Liang, J.-Y, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1994-08-26 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of fructose-1,6-bisphosphatase complexed with fructose 2,6-bisphosphate, AMP, and Zn2+ at 2.0-A resolution: aspects of synergism between inhibitors.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
5OV3
 
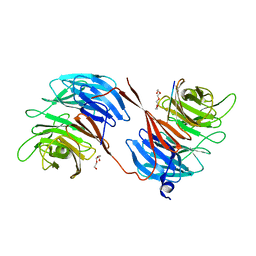 | | Structure of the RbBP5 beta-propeller domain | | Descriptor: | Retinoblastoma-binding protein 5, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Mittal, A, Zhang, Y, Gamblin, S.J, Wilson, J.R. | | Deposit date: | 2017-08-27 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of the RbBP5 beta-propeller domain reveals a surface with potential nucleic acid binding sites.
Nucleic Acids Res., 46, 2018
|
|
7LJC
 
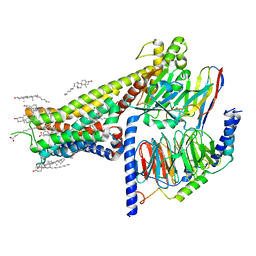 | | Allosteric modulator LY3154207 binding to SKF-81297-bound dopamine receptor 1 in complex with miniGs protein | | Descriptor: | (1R)-6-chloro-1-phenyl-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, ... | | Authors: | Zhuang, Y, Krumm, B, Zhang, H, Zhou, X.E, Wang, Y, Guo, J, Huang, X.-P, Liu, Y, Wang, L, Cheng, X, Jiang, Y, Jiang, H, Melcher, K, Zhang, C, Yi, W, Roth, B.L, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-03 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of dopamine binding and allosteric modulation of the human D1 dopamine receptor.
Cell Res., 31, 2021
|
|
7LJD
 
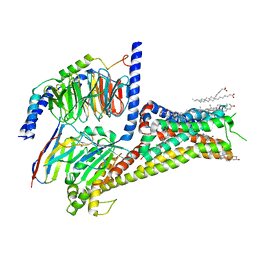 | | Allosteric modulator LY3154207 binding to dopamine-bound dopamine receptor 1 in complex with miniGs protein | | Descriptor: | 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Krumm, B, Zhang, H, Zhou, X.E, Wang, Y, Guo, J, Huang, X.-P, Liu, Y, Wang, L, Cheng, X, Jiang, Y, Jiang, H, Melcher, K, Zhang, C, Yi, W, Roth, B.L, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-03 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of dopamine binding and allosteric modulation of the human D1 dopamine receptor.
Cell Res., 31, 2021
|
|
3J8G
 
 | | Electron cryo-microscopy structure of EngA bound with the 50S ribosomal subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Zhang, X, Yan, K, Zhang, Y, Li, N, Ma, C, Li, Z, Zhang, Y, Feng, B, Liu, J, Sun, Y, Xu, Y, Lei, J, Gao, N. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly
Nucleic Acids Res., 2014
|
|
1FPE
 
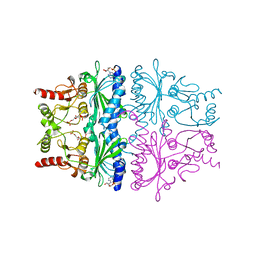 | | STRUCTURAL ASPECTS OF THE ALLOSTERIC INHIBITION OF FRUCTOSE-1,6-BISPHOSPHATASE BY AMP: THE BINDING OF BOTH THE SUBSTRATE ANALOGUE 2,5-ANHYDRO-D-GLUCITOL-1,6-BISPHOSPHATE AND CATALYTIC METAL IONS MONITORED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Villeret, V, Huang, S, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1994-12-15 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural aspects of the allosteric inhibition of fructose-1,6-bisphosphatase by AMP: the binding of both the substrate analogue 2,5-anhydro-D-glucitol 1,6-bisphosphate and catalytic metal ions monitored by X-ray crystallography.
Biochemistry, 34, 1995
|
|
1FPF
 
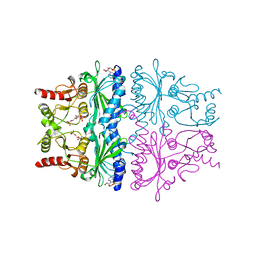 | | STRUCTURAL ASPECTS OF THE ALLOSTERIC INHIBITION OF FRUCTOSE-1,6-BISPHOSPHATASE BY AMP: THE BINDING OF BOTH THE SUBSTRATE ANALOGUE 2,5-ANHYDRO-D-GLUCITOL-1,6-BISPHOSPHATE AND CATALYTIC METAL IONS MONITORED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Villeret, V, Huang, S, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1994-12-15 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural aspects of the allosteric inhibition of fructose-1,6-bisphosphatase by AMP: the binding of both the substrate analogue 2,5-anhydro-D-glucitol 1,6-bisphosphate and catalytic metal ions monitored by X-ray crystallography.
Biochemistry, 34, 1995
|
|
1FPD
 
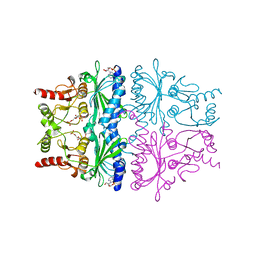 | | STRUCTURAL ASPECTS OF THE ALLOSTERIC INHIBITION OF FRUCTOSE-1,6-BISPHOSPHATASE BY AMP: THE BINDING OF BOTH THE SUBSTRATE ANALOGUE 2,5-ANHYDRO-D-GLUCITOL-1,6-BISPHOSPHATE AND CATALYTIC METAL IONS MONITORED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Villeret, V, Huang, S, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1994-12-15 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural aspects of the allosteric inhibition of fructose-1,6-bisphosphatase by AMP: the binding of both the substrate analogue 2,5-anhydro-D-glucitol 1,6-bisphosphate and catalytic metal ions monitored by X-ray crystallography.
Biochemistry, 34, 1995
|
|
4Y5T
 
 | | Structure of FtmOx1 apo with metal Iron | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COBALT (II) ION, FE (II) ION, ... | | Authors: | Yan, W, Zhang, Y. | | Deposit date: | 2015-02-12 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Endoperoxide formation by an alpha-ketoglutarate-dependent mononuclear non-haem iron enzyme.
Nature, 527, 2015
|
|
4Y5S
 
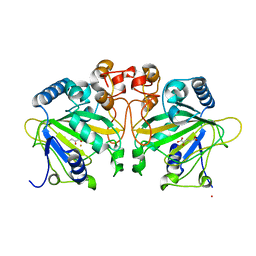 | | Structure of FtmOx1 with a-Ketoglutarate as co-substrate | | Descriptor: | 2-OXOGLUTARIC ACID, COBALT (II) ION, FE (II) ION, ... | | Authors: | Yan, W, Zhang, Y. | | Deposit date: | 2015-02-12 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Endoperoxide formation by an alpha-ketoglutarate-dependent mononuclear non-haem iron enzyme.
Nature, 527, 2015
|
|
1FPG
 
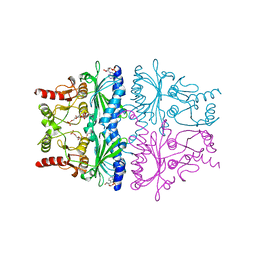 | | STRUCTURAL ASPECTS OF THE ALLOSTERIC INHIBITION OF FRUCTOSE-1,6-BISPHOSPHATASE BY AMP: THE BINDING OF BOTH THE SUBSTRATE ANALOGUE 2,5-ANHYDRO-D-GLUCITOL-1,6-BISPHOSPHATE AND CATALYTIC METAL IONS MONITORED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Villeret, V, Huang, S, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1994-12-15 | | Release date: | 1995-02-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural aspects of the allosteric inhibition of fructose-1,6-bisphosphatase by AMP: the binding of both the substrate analogue 2,5-anhydro-D-glucitol 1,6-bisphosphate and catalytic metal ions monitored by X-ray crystallography.
Biochemistry, 34, 1995
|
|
1P58
 
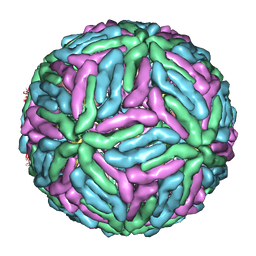 | | Complex Organization of Dengue Virus Membrane Proteins as Revealed by 9.5 Angstrom Cryo-EM reconstruction | | Descriptor: | Envelope protein M, Major envelope protein E | | Authors: | Zhang, W, Chipman, P.R, Corver, J, Johnson, P.R, Zhang, Y, Mukhopadhyay, S, Baker, T.S, Strauss, J.H, Rossmann, M.G, Kuhn, R.J. | | Deposit date: | 2003-04-25 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Visualization of membrane protein domains by cryo-electron microscopy of dengue virus
Nat.Struct.Biol., 10, 2003
|
|
2RFS
 
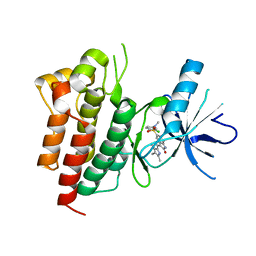 | | X-ray structure of SU11274 bound to c-Met | | Descriptor: | Hepatocyte growth factor receptor, N-(3-chlorophenyl)-N-methyl-2-oxo-3-[(3,4,5-trimethyl-1H-pyrrol-2-yl)methyl]-2H-indole-5-sulfonamide | | Authors: | Bellon, S.F, Kaplan-Lefko, P, Yang, Y, Zhang, Y, Moriguchi, J, Dussault, I. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | c-Met inhibitors with novel binding mode show activity against several hereditary papillary renal cell carcinoma-related mutations.
J.Biol.Chem., 283, 2008
|
|
7CZ5
 
 | | Cryo-EM structure of the human growth hormone-releasing hormone receptor-Gs protein complex | | Descriptor: | CHOLESTEROL, Growth hormone-releasing hormone receptor,growth hormone-releasing hormone receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhou, F, Zhang, H, Cong, Z, Zhao, L, Zhou, Q, Mao, C, Cheng, X, Shen, D, Cai, X, Ma, C, Wang, Y, Dai, A, Zhou, Y, Sun, W, Zhao, F, Zhao, S, Jiang, H, Jiang, Y, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2020-09-07 | | Release date: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for activation of the growth hormone-releasing hormone receptor.
Nat Commun, 11, 2020
|
|
7D68
 
 | | Cryo-EM structure of the human glucagon-like peptide-2 receptor-Gs protein complex | | Descriptor: | Glucagon-like peptide 2 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, W, Chen, L, Zhou, Q, Zhao, L, Zhang, H, Cong, Z, Shen, D, Zhao, F, Zhou, F, Cai, X, Chen, Y, Zhou, Y, Gadgaard, S, van der Velden, W.J, Zhao, S, Jiang, Y, Rosenkilde, M.M, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A unique hormonal recognition feature of the human glucagon-like peptide-2 receptor.
Cell Res., 30, 2020
|
|
2RFN
 
 | | x-ray structure of c-Met with inhibitor. | | Descriptor: | 2-benzyl-5-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-3-methylpyrimidin-4(3H)-one, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Kaplan-Lefko, P, Yang, Y, Zhang, Y, Moriguchi, J, Dussault, I. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | c-Met inhibitors with novel binding mode show activity against several hereditary papillary renal cell carcinoma-related mutations.
J.Biol.Chem., 283, 2008
|
|
6PWC
 
 | | A complex structure of arrestin-2 bound to neurotensin receptor 1 | | Descriptor: | Beta-arrestin-1, Fab30 heavy chain, Fab30 light chain, ... | | Authors: | Yin, W, Li, Z, Jin, M, Yin, Y.-L, de Waal, P.W, Pal, K, Gao, X, He, Y, Gao, J, Wang, X, Zhang, Y, Zhou, H, Melcher, K, Jiang, Y, Cong, Y, Zhou, X.E, Yu, X, Xu, H.E. | | Deposit date: | 2019-07-22 | | Release date: | 2019-12-04 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | A complex structure of arrestin-2 bound to a G protein-coupled receptor.
Cell Res., 29, 2019
|
|
4ZA1
 
 | | Crystal Structure of NosA Involved in Nosiheptide Biosynthesis | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NosA | | Authors: | Liu, S, Guo, H, Zhang, T, Han, L, Yao, P, Zhang, Y, Rong, N, Yu, Y, Lan, W, Wang, C, Ding, J, Wang, R, Liu, W, Cao, C. | | Deposit date: | 2015-04-13 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based Mechanistic Insights into Terminal Amide Synthase in Nosiheptide-Represented Thiopeptides Biosynthesis
Sci Rep, 5, 2015
|
|
