5X2G
 
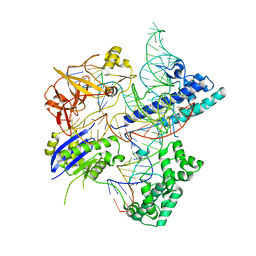 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
5X2H
 
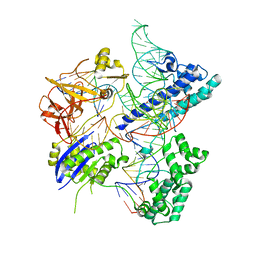 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
1GDV
 
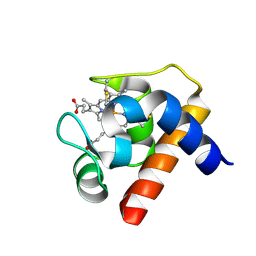 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 FROM RED ALGA PORPHYRA YEZOENSIS AT 1.57 A RESOLUTION | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Yamada, S, Park, S.-Y, Shimizu, H, Shiro, Y, Oku, T. | | Deposit date: | 2000-10-06 | | Release date: | 2001-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure of cytochrome c6 from the red alga Porphyra yezoensis at 1. 57 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
7XHT
 
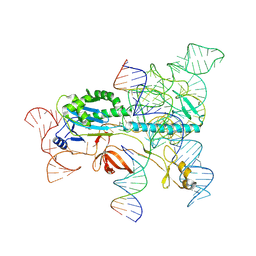 | | Structure of the OgeuIscB-omega RNA-target DNA complex | | Descriptor: | DNA (49-MER), DNA (5'-D(P*GP*AP*AP*GP*AP*AP*AP*AP*CP*CP*AP*T)-3'), LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Kato, K, Okazaki, O, Isayama, Y, Ishikawa, J, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-04-10 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structure of the IscB-omega RNA ribonucleoprotein complex, the likely ancestor of CRISPR-Cas9.
Nat Commun, 13, 2022
|
|
7V5N
 
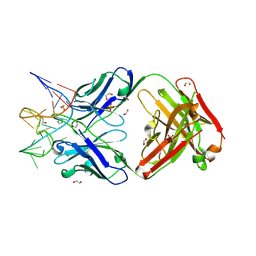 | | Crystal structure of Fab fragment of bevacizumab bound to DNA aptamer | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*GP*TP*TP*GP*GP*TP*GP*GP*TP*AP*GP*TP*TP*AP*CP*GP*TP*TP*CP*GP*C)-3'), IMIDAZOLE, ... | | Authors: | Hishiki, A, Tong, J, Todoroki, K, Hashimoto, H. | | Deposit date: | 2021-08-17 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of a DNA aptamer that binds to the complementarity-determining region of therapeutic monoclonal antibody and affinity improvement induced by pH-change for sensitive detection.
Biosens.Bioelectron., 203, 2022
|
|
7V6B
 
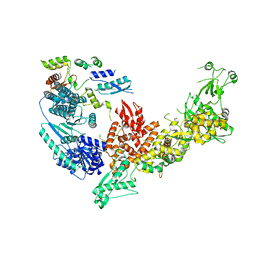 | | Structure of the Dicer-2-R2D2 heterodimer | | Descriptor: | Dicer-2, isoform A, R2D2 | | Authors: | Yamaguchi, S, Nishizawa, T, Kusakizako, T, Yamashita, K, Tomita, A, Hirano, H, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-20 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex.
Nature, 607, 2022
|
|
7V6C
 
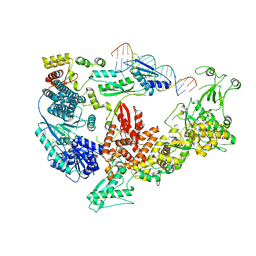 | | Structure of the Dicer-2-R2D2 heterodimer bound to small RNA duplex | | Descriptor: | Dicer-2, isoform A, R2D2, ... | | Authors: | Yamaguchi, S, Nishizawa, T, Kusakizako, T, Yamashita, K, Tomita, A, Hirano, H, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-20 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex.
Nature, 607, 2022
|
|
5YY3
 
 | |
5YY2
 
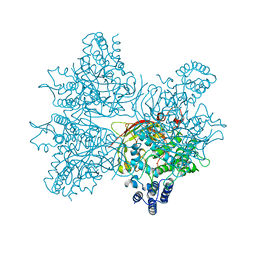 | | Crystal structure of AsqI with Zn | | Descriptor: | Uncharacterized protein AsqI, ZINC ION | | Authors: | Hara, K, Hashimoto, H, Kishimoto, S, Watanabe, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Enzymatic one-step ring contraction for quinolone biosynthesis.
Nat Commun, 9, 2018
|
|
2AI5
 
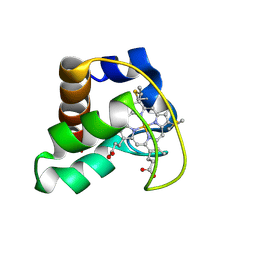 | | Solution Structure of Cytochrome C552, determined by Distributed Computing Implementation for NMR data | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Nakamura, S, Ichiki, S.I, Takashima, H, Uchiyama, S, Hasegawa, J, Kobayashi, Y, Sambongi, Y, Ohkubo, T. | | Deposit date: | 2005-07-29 | | Release date: | 2006-05-23 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure of Cytochrome c552 from a Moderate Thermophilic Bacterium, Hydrogenophilus thermoluteolus: Comparative Study on the Thermostability of Cytochrome c
Biochemistry, 45, 2006
|
|
7CP7
 
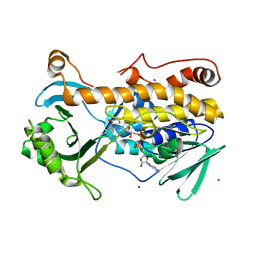 | | Crystal structure of FqzB, native proteins | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IODIDE ION, MAK1-like monooxygenase | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2020-08-06 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Analyses of a Spiro-Carbon-Forming, Highly Promiscuous Epoxidase from Fungal Natural Product Biosynthesis.
Biochemistry, 59, 2020
|
|
7C7L
 
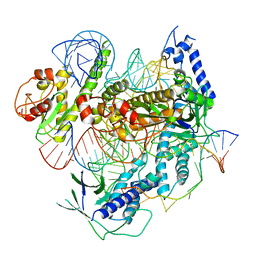 | | Cryo-EM structure of the Cas12f1-sgRNA-target DNA complex | | Descriptor: | CRISPR-associated protein Cas14a.1, DNA (40-mer), ZINC ION, ... | | Authors: | Takeda, N.S, Nakagawa, R, Okazaki, S, Hirano, H, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2020-05-26 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the miniature type V-F CRISPR-Cas effector enzyme.
Mol.Cell, 81, 2021
|
|
5B2T
 
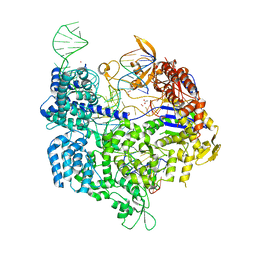 | | Crystal structure of the Streptococcus pyogenes Cas9 VRER variant in complex with sgRNA and target DNA (TGCG PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CRISPR-associated endonuclease Cas9, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9
Mol.Cell, 61, 2016
|
|
5B2S
 
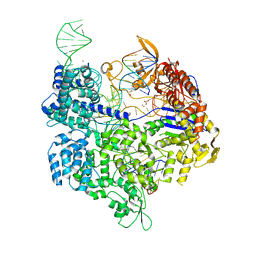 | | Crystal structure of the Streptococcus pyogenes Cas9 EQR variant in complex with sgRNA and target DNA (TGAG PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CRISPR-associated endonuclease Cas9, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9
Mol.Cell, 61, 2016
|
|
5B2R
 
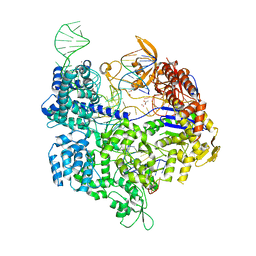 | | Crystal structure of the Streptococcus pyogenes Cas9 VQR variant in complex with sgRNA and target DNA (TGA PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CRISPR-associated endonuclease Cas9, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9
Mol.Cell, 61, 2016
|
|
7CP6
 
 | | Crystal structure of FqzB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IODIDE ION, MAK1-like monooxygenase, ... | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2020-08-06 | | Release date: | 2020-12-30 | | Last modified: | 2021-01-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analyses of a Spiro-Carbon-Forming, Highly Promiscuous Epoxidase from Fungal Natural Product Biosynthesis.
Biochemistry, 59, 2020
|
|
7FI3
 
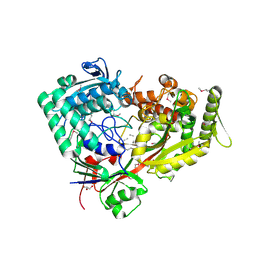 | | Archaeal oligopeptide permease A (OppA) from Thermococcus kodakaraensis in complex with an endogenous pentapeptide | | Descriptor: | ABC-type dipeptide/oligopeptide transport system, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Yokoyama, H, Kamei, N, Konishi, K, Hara, K, Hashimoto, H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-04-13 | | Last modified: | 2022-06-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for peptide recognition by archaeal oligopeptide permease A.
Proteins, 90, 2022
|
|
6IRZ
 
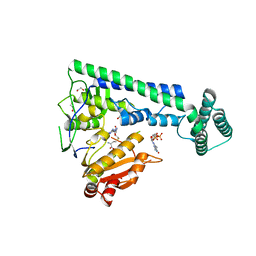 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | Descriptor: | 1,2-ETHANEDIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, PDX1 C-terminal-inhibiting factor 1, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IRV
 
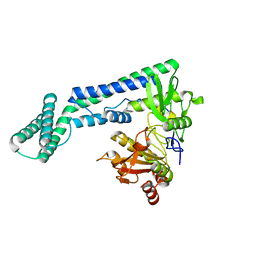 | |
6IS0
 
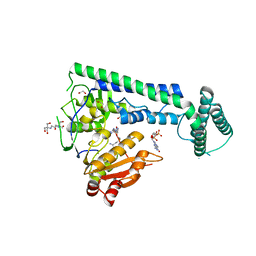 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IRY
 
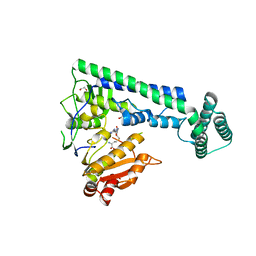 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH | | Descriptor: | 1,2-ETHANEDIOL, PDX1 C-terminal-inhibiting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IRX
 
 | |
1RG5
 
 | | Structure of the photosynthetic reaction centre from Rhodobacter sphaeroides carotenoidless strain R-26.1 | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Roszak, A.W, Hashimoto, H, Gardiner, A.T, Cogdell, R.J, Isaacs, N.W. | | Deposit date: | 2003-11-11 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Regulation of Carotenoid Binding: Gatekeeper and Locking Amino Acid Residues in Reaction Centers of Rhodobacter sphaeroides
STRUCTURE, 12, 2004
|
|
6JOO
 
 | | Crystal structure of Corynebacterium diphtheriae Cas9 in complex with sgRNA and target DNA | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated protein,CRISPR-associated endonuclease Cas9, Guide RNA, ... | | Authors: | Hirano, S, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2019-03-22 | | Release date: | 2019-04-17 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the promiscuous PAM recognition by Corynebacterium diphtheriae Cas9.
Nat Commun, 10, 2019
|
|
1RQK
 
 | | Structure of the reaction centre from Rhodobacter sphaeroides carotenoidless strain R-26.1 reconstituted with 3,4-dihydrospheroidene | | Descriptor: | 3,4-DIHYDROSPHEROIDENE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Roszak, A.W, Hashimoto, H, Gardiner, A.T, Cogdell, R.J, Isaacs, N.W. | | Deposit date: | 2003-12-05 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein Regulation of Carotenoid Binding: Gatekeeper and Locking Amino Acid Residues in Reaction Centers of Rhodobacter sphaeroides
STRUCTURE, 12, 2004
|
|
