4OJ3
 
 | |
4OJG
 
 | |
3IRQ
 
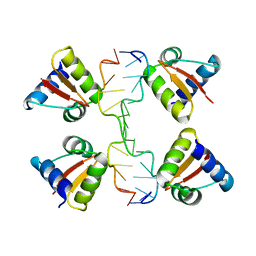 | | Crystal structure of a Z-Z junction | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*CP*GP*CP*GP*AP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*TP*CP*GP*CP*GP*CP*G)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Athanasiadis, A, de Rosa, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a junction between two Z-DNA helices.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IRR
 
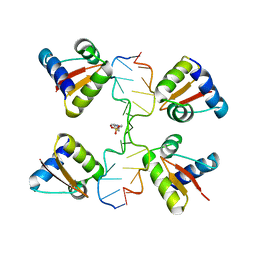 | | Crystal Structure of a Z-Z junction (with HEPES intercalating) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA (5'-D(*A*CP*CP*GP*CP*GP*CP*GP*AP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*G*TP*CP*GP*CP*GP*CP*GP*TP*CP*GP*CP*GP*CP*G)-3'), ... | | Authors: | Athanasiadis, A, de Rosa, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of a junction between two Z-DNA helices.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6Q9Z
 
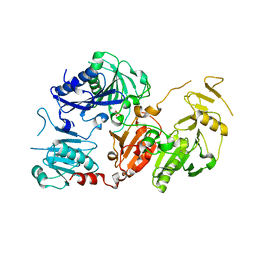 | | Crystal structure of the pathological G167R variant of calcium-free human gelsolin, | | Descriptor: | GLYCEROL, Gelsolin, SULFATE ION | | Authors: | Boni, F, Scalone, E, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
6QBF
 
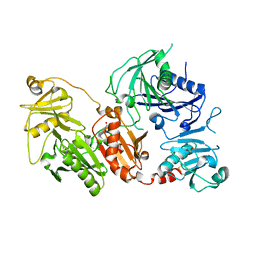 | | Crystal structure of the pathological D187N variant of calcium-free human gelsolin. | | Descriptor: | GLYCEROL, Gelsolin, SODIUM ION, ... | | Authors: | Scalone, E, Boni, F, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-21 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.499 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
6Q9R
 
 | | Crystal structure of the pathological N184K variant of calcium-free human gelsolin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Scalone, E, Boni, F, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
2V2K
 
 | | THE CRYSTAL STRUCTURE OF FDXA, A 7FE FERREDOXIN FROM MYCOBACTERIUM SMEGMATIS | | Descriptor: | ACETATE ION, FE3-S4 CLUSTER, FERREDOXIN | | Authors: | Ricagno, S, de Rosa, M, Aliverti, A, Zanetti, G, Bolognesi, M. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Fdxa, a 7Fe Ferredoxin from Mycobacterium Smegmatis.
Biochem.Biophys.Res.Commun., 360, 2007
|
|
7P2B
 
 | |
4BND
 
 | | Structure of an atypical alpha-phosphoglucomutase similar to eukaryotic phosphomannomutases | | Descriptor: | ALPHA-PHOSPHOGLUCOMUTASE, GLYCEROL, SULFATE ION | | Authors: | Nogly, P, Matias, P.M, De Rosa, M, Castro, R, Santos, H, Neves, A.R, Archer, M. | | Deposit date: | 2013-05-14 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-Resolution Structure of an Atypical [Alpha]-Phosphoglucomutase Related to Eukaryotic Phosphomannomutases
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5FAE
 
 | | N184K pathological variant of gelsolin domain 2 (trigonal form) | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boni, F, Milani, M, Ricagno, S, Bolognesi, M, de Rosa, M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of a novel renal amyloidosis due to N184K gelsolin variant.
Sci Rep, 6, 2016
|
|
3DHM
 
 | | Beta 2 microglobulin mutant D59P | | Descriptor: | Beta-2-microglobulin | | Authors: | Ricagno, S, Colombo, M, de Rosa, M, Bolognesi, M, Giorgetti, S, Bellotti, V. | | Deposit date: | 2008-06-18 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DE loop mutations affect beta2-microglobulin stability and amyloid aggregation
Biochem.Biophys.Res.Commun., 377, 2008
|
|
5FAF
 
 | | N184K pathological variant of gelsolin domain 2 (orthorhombic form) | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Boni, F, Milani, M, Ricagno, s, Bolognesi, M, de Rosa, M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Molecular basis of a novel renal amyloidosis due to N184K gelsolin variant.
Sci Rep, 6, 2016
|
|
5O2Z
 
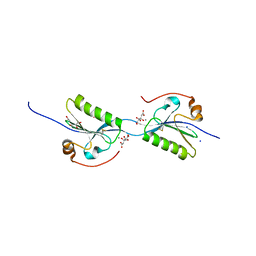 | | Domain swap dimer of the G167R variant of gelsolin second domain | | Descriptor: | ACETATE ION, CALCIUM ION, CITRATE ANION, ... | | Authors: | Boni, F, Milani, M, Mastrangelo, E, de Rosa, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Gelsolin pathogenic Gly167Arg mutation promotes domain-swap dimerization of the protein.
Hum. Mol. Genet., 27, 2018
|
|
3DHJ
 
 | | Beta 2 microglobulin mutant W60C | | Descriptor: | Beta-2-microglobulin | | Authors: | Ricagno, S, Colombo, M, de Rosa, M, Bolognesi, M, Giorgetti, S, Bellotti, V. | | Deposit date: | 2008-06-18 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DE loop mutations affect beta2-microglobulin stability and amyloid aggregation
Biochem.Biophys.Res.Commun., 377, 2008
|
|
5CKA
 
 | | Human beta-2 microglobulin double mutant W60G-N83V | | Descriptor: | ACETATE ION, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sala, B.M, De Rosa, M, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-15 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational design of mutations that change the aggregation rate of a protein while maintaining its native structure and stability.
Sci Rep, 6, 2016
|
|
5CKG
 
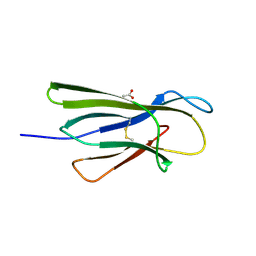 | | Human beta-2 microglobulin mutant V85E | | Descriptor: | ACETATE ION, Beta-2-microglobulin, GLYCEROL | | Authors: | Sala, B.M, De Rosa, M, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-15 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rational design of mutations that change the aggregation rate of a protein while maintaining its native structure and stability.
Sci Rep, 6, 2016
|
|
5CFH
 
 | |
5CSG
 
 | |
5CSB
 
 | | The crystal structure of beta2-microglobulin D76N mutant at room temperature | | Descriptor: | Beta-2-microglobulin | | Authors: | de Rosa, M, Mota, C.S, de Sanctis, D, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity.
Nat Commun, 9, 2018
|
|
5CS7
 
 | | The crystal structure of wt beta2-microglobulin at room temperature | | Descriptor: | Beta-2-microglobulin | | Authors: | de Rosa, M, Mota, C.S, de Sanctis, D, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity.
Nat Commun, 9, 2018
|
|
4RMW
 
 | |
4RMU
 
 | |
4RMV
 
 | |
2C7G
 
 | | FprA from Mycobacterium tuberculosis: His57Gln mutant | | Descriptor: | 4-OXO-NICOTINAMIDE-ADENINE DINUCLEOTIDE PHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH-FERREDOXIN REDUCTASE FPRA, ... | | Authors: | Pennati, A, Razeto, A, De Rosa, M, Pandini, V, Vanoni, M.A, Aliverti, A, Mattevi, A, Coda, A, Zanetti, G. | | Deposit date: | 2005-11-24 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the His57-Glu214 Ionic Couple Located in the Active Site of Mycobacterium Tuberculosis Fpra.
Biochemistry, 45, 2006
|
|
