5KW2
 
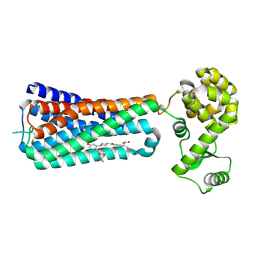 | | The extra-helical binding site of GPR40 and the structural basis for allosteric agonism and incretin stimulation | | Descriptor: | (3~{S})-3-cyclopropyl-3-[2-[1-[2-[2,2-dimethylpropyl-(6-methylpyridin-2-yl)carbamoyl]-5-methoxy-phenyl]piperidin-4-yl]-1-benzofuran-6-yl]propanoic acid, Free fatty acid receptor 1,Lysozyme,Free fatty acid receptor 1 | | Authors: | Ho, J.D, Chau, B, Rodgers, L, Lu, F, Wilbur, K.L, Otto, K.A, Chen, Y, Song, M, Riley, J.P, Yang, H.-C, Reynolds, N.A, Kahl, S.D, Lewis, A.P, Groshong, C, Madsen, R.E, Conners, K, Linswala, J.P, Gheyi, T, Saflor, M.D, Lee, M.R, Benach, J, Baker, K.A, Montrose-Rafizadeh, C, Genin, M.J, Miller, A.R, Hamdouchi, C. | | Deposit date: | 2016-07-15 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural basis for GPR40 allosteric agonism and incretin stimulation.
Nat Commun, 9, 2018
|
|
5ZFX
 
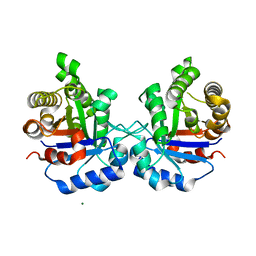 | | Crystal Structure of Triosephosphate isomerase from Opisthorchis viverrini | | Descriptor: | MAGNESIUM ION, Triosephosphate isomerase | | Authors: | Son, J, Kim, S, Kim, S.E, Lee, H, Lee, M.R, Hwang, K.Y. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Structural Analysis of an Epitope Candidate of Triosephosphate Isomerase in Opisthorchis viverrini.
Sci Rep, 8, 2018
|
|
5ZG5
 
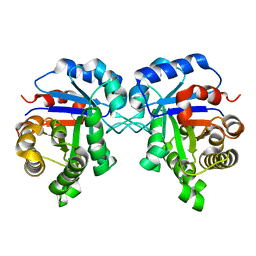 | | Crystal Structure of Triosephosphate isomerase SADsubAAA mutant from Opisthorchis viverrini | | Descriptor: | Triosephosphate isomerase | | Authors: | Son, J, Kim, S, Kim, S.E, Lee, H, Lee, M.R, Hwang, K.Y. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Structural Analysis of an Epitope Candidate of Triosephosphate Isomerase in Opisthorchis viverrini.
Sci Rep, 8, 2018
|
|
5ZGA
 
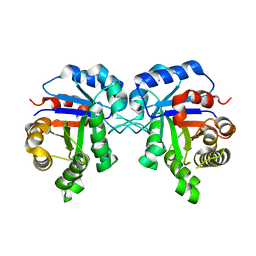 | | Crystal Structure of Triosephosphate isomerase SAD deletion and N115A mutant from Opisthorchis viverrini | | Descriptor: | Triosephosphate isomerase | | Authors: | Son, J, Kim, S, Kim, S.E, Lee, H, Lee, M.R, Hwang, K.Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Structural Analysis of an Epitope Candidate of Triosephosphate Isomerase in Opisthorchis viverrini.
Sci Rep, 8, 2018
|
|
5ZG4
 
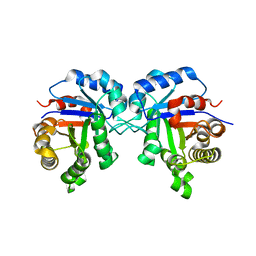 | | Crystal Structure of Triosephosphate isomerase SAD deletion mutant from Opisthorchis viverrini | | Descriptor: | Triosephosphate isomerase | | Authors: | Son, J, Kim, S, Kim, S.E, Lee, H, Lee, M.R, Hwang, K.Y. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Structural Analysis of an Epitope Candidate of Triosephosphate Isomerase in Opisthorchis viverrini.
Sci Rep, 8, 2018
|
|
6VGI
 
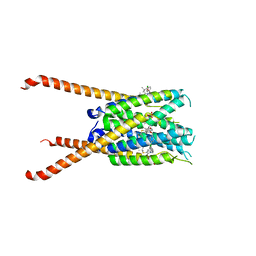 | | Crystal Structures of FLAP bound to MK-866 | | Descriptor: | 3-[3-(tert-butylsulfanyl)-1-[(4-chlorophenyl)methyl]-5-(propan-2-yl)-1H-indol-2-yl]-2,2-dimethylpropanoic acid, 5-lipoxygenase-activating protein, SULFATE ION | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6VGC
 
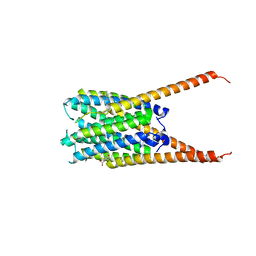 | | Crystal Structures of FLAP bound to DG-031 | | Descriptor: | (2R)-cyclopentyl{4-[(quinolin-2-yl)methoxy]phenyl}acetic acid, 5-lipoxygenase-activating protein, CALCIUM ION, ... | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6VL4
 
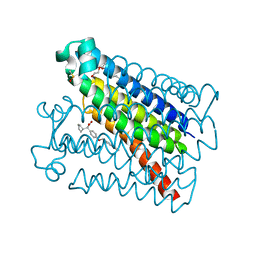 | | Crystal Structure of mPGES-1 bound to DG-031 | | Descriptor: | (2R)-cyclopentyl{4-[(quinolin-2-yl)methoxy]phenyl}acetic acid, Prostaglandin E synthase, TETRAETHYLENE GLYCOL, ... | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-22 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
1YFM
 
 | |
3U8W
 
 | | Crystal Structure of p38a Mitogen-Activated Protein Kinase in Complex with a Triazolopyridazinone inhibitor | | Descriptor: | 3-[3-(2-chloro-6-fluorophenyl)-5-ethyl-6-oxo-5,6-dihydro[1,2,4]triazolo[4,3-b]pyridazin-7-yl]-N-cyclopropyl-4-methylbenzamide, Mitogen-activated protein kinase 14 | | Authors: | Mohr, C, Jordan, S. | | Deposit date: | 2011-10-17 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of triazolopyridazinones as potent p38alpha inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5KZI
 
 | | Crystal structure of human Pim-1 kinase in complex with an imidazopyridazine inhibitor. | | Descriptor: | Serine/threonine-protein kinase pim-1, ~{N}-[4-[(3~{S})-3-azanylpiperidin-1-yl]pyridin-3-yl]-2-[2,6-bis(fluoranyl)phenyl]imidazo[1,5-b]pyridazin-7-amine | | Authors: | Mohr, C. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of imidazopyridazines as potent Pim-1/2 kinase inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
4TY1
 
 | |
3L9M
 
 | | Crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 18 | | Descriptor: | (2S)-N~1~-[5-(3-methyl-1H-indazol-5-yl)-1,3,4-thiadiazol-2-yl]-3-(4-methylphenyl)propane-1,2-diamine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LHJ
 
 | | Crystal Structure of p38a Mitogen-Activated Protein Kinase in Complex with a Pyrazolopyridinone Inhibitor. | | Descriptor: | Mitogen-activated protein kinase 14, N-cyclopropyl-3-[1-(2,4-difluorophenyl)-7-methyl-6-oxo-6,7-dihydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-4-methylbenzamide | | Authors: | Mohr, C, Jordan, S. | | Deposit date: | 2010-01-22 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Discovery and evaluation of 7-alkyl-1,5-bis-aryl-pyrazolopyridinones as highly potent, selective, and orally efficacious inhibitors of p38alpha mitogen-activated protein kinase.
J.Med.Chem., 53, 2010
|
|
7XDG
 
 | | Cryo-EM structures of human mitochondrial NAD(P)+-dependent malic enzyme in a ternary complex with NAD+ and allosteric inhibitor MDSA | | Descriptor: | 5-[(3-carboxy-4-oxidanyl-phenyl)methyl]-2-oxidanyl-benzoic acid, NAD-dependent malic enzyme, mitochondrial, ... | | Authors: | Wang, C.H, Hsieh, J.T, Ho, M.C, Hung, H.C. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Suppression of the human malic enzyme 2 modifies energy metabolism and inhibits cellular respiration
Commun Biol, 6, 2023
|
|
7XDF
 
 | | Cryo-EM structures of human mitochondrial NAD(P)+-dependent malic enzyme in a ternary complex with NAD+ and allosteric inhibitor EA | | Descriptor: | 4-[(3-carboxy-2-oxidanyl-naphthalen-1-yl)methyl]-3-oxidanyl-naphthalene-2-carboxylic acid, NAD-dependent malic enzyme, mitochondrial, ... | | Authors: | Wang, C.H, Hsieh, J.T, Ho, M.C, Hung, H.C. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Suppression of the human malic enzyme 2 modifies energy metabolism and inhibits cellular respiration
Commun Biol, 6, 2023
|
|
7XDE
 
 | | Cryo-EM structures of human mitochondrial NAD(P)+-dependent malic enzyme in apo form | | Descriptor: | NAD-dependent malic enzyme, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, C.H, Hsieh, J.T, Ho, M.C, Hung, H.C. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Suppression of the human malic enzyme 2 modifies energy metabolism and inhibits cellular respiration
Commun Biol, 6, 2023
|
|
7P6N
 
 | | ROCK2 IN COMPLEX WITH COMPOUND 12 | | Descriptor: | Rho-associated protein kinase 2, ~{N}-[(1~{R})-1-(3-methoxyphenyl)ethyl]-4-pyridin-4-yl-piperidine-1-carboxamide | | Authors: | Maillard, M.C. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of a Potent, Selective, and Brain-Penetrant Rho Kinase Inhibitor and its Activity in a Mouse Model of Huntington's Disease.
J.Med.Chem., 65, 2022
|
|
5X2D
 
 | |
5EOL
 
 | |
5X2E
 
 | |
5FD2
 
 | |
4YK5
 
 | | Crystal Structures of mPGES-1 Inhibitor Complexes | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3-[1-(4-chlorobenzyl)-5-(2-fluoro-2'-methylbiphenyl-4-yl)-3-methyl-1H-indol-2-yl]-2,2-dimethylpropanoic acid, GLUTATHIONE, ... | | Authors: | Luz, J.G, Antonysamy, S, Kuklish, S.L, Fisher, M.J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structures of mPGES-1 Inhibitor Complexes Form a Basis for the Rational Design of Potent Analgesic and Anti-Inflammatory Therapeutics.
J.Med.Chem., 58, 2015
|
|
4YL1
 
 | | Crystal Structures of mPGES-1 Inhibitor Complexes | | Descriptor: | 5-(4-tert-butylphenyl)-1-[4-(propan-2-yloxy)phenyl]-1H-indole-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Luz, J.G, Antonysamy, S, Kuklish, S.L, Fisher, M.J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structures of mPGES-1 Inhibitor Complexes Form a Basis for the Rational Design of Potent Analgesic and Anti-Inflammatory Therapeutics.
J.Med.Chem., 58, 2015
|
|
4WRS
 
 | |
