5MW4
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD7 [N-(3-(((R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl)(methyl)amino)propyl)-2-(3-(2-chloro-3-(2-methylpyridin-3-yl)benzo[b]thiophen-5-yl)ureido)acetamide] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N~2~-{[2-chloro-3-(2-methylpyridin-3-yl)-1-benzothiophen-5-yl]carbamoyl}-N-(3-{methyl[(3R)-1-(5H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]amino}propyl)glycinamide, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
5MW3
 
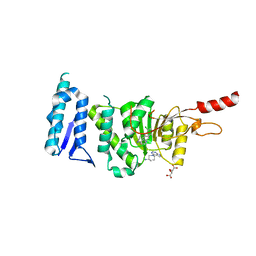 | | Crystal structure of Dot1L in complex with inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] and inhibitor CPD2 [(R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine] | | Descriptor: | (3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
5MVS
 
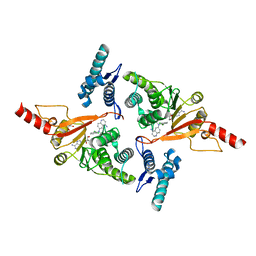 | | Crystal structure of Dot1L in complex with adenosine and inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] | | Descriptor: | ADENOSINE, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-17 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
5DT2
 
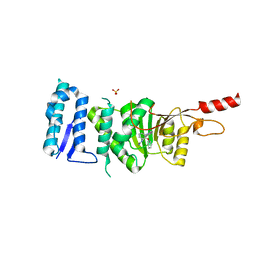 | | Crystal structure of Dot1L in complex with inhibitor CPD11 [N4-methyl-N2-(2-methyl-1-(2-phenoxyphenyl)-1H-indol-6-yl)pyrimidine-2,4-diamine] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N~4~-methyl-N~2~-[2-methyl-1-(2-phenoxyphenyl)-1H-indol-6-yl]pyrimidine-2,4-diamine, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-17 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
5DRY
 
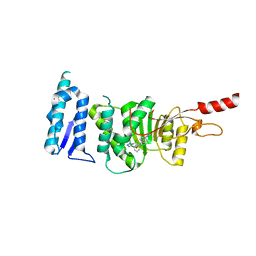 | | Crystal structure of Dot1L in complex with inhibitor CPD3 [N-(1-(2-chlorophenyl)-1H-indol-6-yl)-2-(2-(5-(2-chlorophenyl)-1H-tetrazol-1-yl)acetyl)hydrazinecarboxamide] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N-[1-(2-chlorophenyl)-1H-indol-6-yl]-2-{[5-(2-chlorophenyl)-1H-tetrazol-1-yl]acetyl}hydrazinecarboxamide, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-16 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
5DSX
 
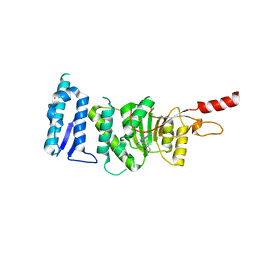 | | Crystal structure of Dot1L in complex with inhibitor CPD10 [6'-chloro-1,4-dimethyl-5'-(2-methyl-6-((4-(methylamino)pyrimidin-2-yl)amino)-1H-indol-1-yl)-[3,3'-bipyridin]-2(1H)-one] | | Descriptor: | 6'-chloro-1,4-dimethyl-5'-(2-methyl-6-{[4-(methylamino)pyrimidin-2-yl]amino}-1H-indol-1-yl)-3,3'-bipyridin-2(1H)-one, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-17 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
5DRT
 
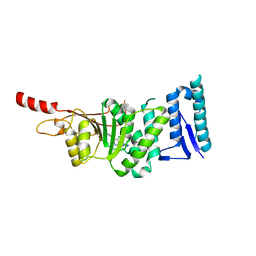 | | Crystal structure of Dot1L in complex with inhibitor CPD2 [2-(2-(5-((2-chlorophenoxy)methyl)-1H-tetrazol-1-yl)acetyl)-N-(4-chlorophenyl)hydrazinecarboxamide] | | Descriptor: | 2-({5-[(2-chlorophenoxy)methyl]-1H-tetrazol-1-yl}acetyl)-N-(4-chlorophenyl)hydrazinecarboxamide, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-16 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
3ZFJ
 
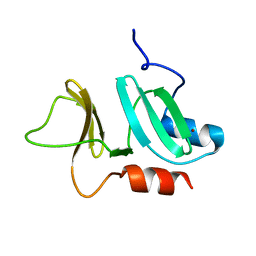 | | N-terminal domain of pneumococcal PhtD protein with bound Zn(II) | | Descriptor: | PNEUMOCOCCAL HISTIDINE TRIAD PROTEIN D, ZINC ION | | Authors: | Bersch, B, Bougault, C, Favier, A, Gabel, F, Roux, L, Vernet, T, Durmort, C. | | Deposit date: | 2012-12-11 | | Release date: | 2013-11-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | New Insights Into Histidine Triad Proteins: Solution Structure of a Streptococcus Pneumoniae Phtd Domain and Zinc Transfer to Adcaii.
Plos One, 8, 2013
|
|
3ZQD
 
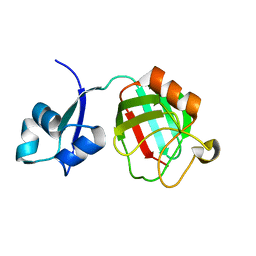 | | B. subtilis L,D-transpeptidase | | Descriptor: | L, D-TRANSPEPTIDASE YKUD | | Authors: | Lecoq, L, Simorre, J.-P, Bougault, C, Arthur, M, Hugonnet, J.-E, Veckerle, C, Pessey, O. | | Deposit date: | 2011-06-09 | | Release date: | 2012-05-23 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Dynamics Induced by Beta-Lactam Antibiotics in the Active Site of Bacillus subtilis L,D-Transpeptidase.
Structure, 20, 2012
|
|
3ZG4
 
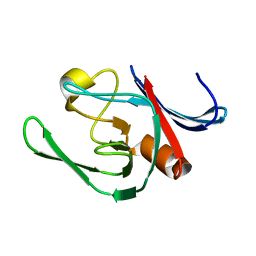 | | NMR structure of the catalytic domain from E. faecium L,D- transpeptidase | | Descriptor: | ERFK/YBIS/YCFS/YNHG | | Authors: | Lecoq, L, Dubee, V, Triboulet, S, Bougault, C, Hugonnet, J.E, Arthur, M, Simorre, J.P. | | Deposit date: | 2012-12-14 | | Release date: | 2013-04-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structure of Enterococcus Faecium L,D---Transpeptidase Acylated by Ertapenem Provides Insight Into the Inactivation Mechanism.
Acs Chem.Biol., 8, 2013
|
|
4A52
 
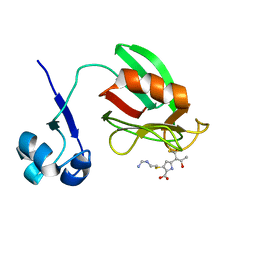 | | NMR structure of the imipenem-acylated L,D-transpeptidase from Bacillus subtilis | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, PUTATIVE L, D-TRANSPEPTIDASE YKUD | | Authors: | Lecoq, L, Simorre, J, Bougault, C, Arthur, M, Hugonnet, J, Veckerle, C, Pessey, O. | | Deposit date: | 2011-10-24 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Dynamics Induced by Beta-Lactam Antibiotics in the Active Site of Bacillus subtilis L,D-Transpeptidase.
Structure, 20, 2012
|
|
3ZGP
 
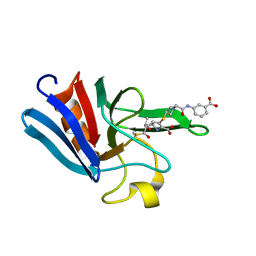 | | NMR structure of the catalytic domain from E. faecium L,D- transpeptidase acylated by ertapenem | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, ERFK/YBIS/YCFS/YNHG | | Authors: | Lecoq, L, Triboulet, S, Dubee, V, Bougault, C, Hugonnet, J.E, Arthur, M, Simorre, J.P. | | Deposit date: | 2012-12-18 | | Release date: | 2013-04-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Structure of Enterococcus Faecium L,D---Transpeptidase Acylated by Ertapenem Provides Insight Into the Inactivation Mechanism.
Acs Chem.Biol., 8, 2013
|
|
2MHK
 
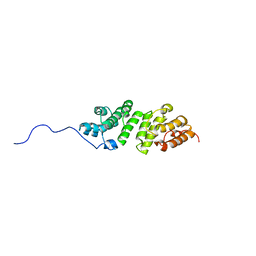 | | E. coli LpoA N-terminal domain | | Descriptor: | Penicillin-binding protein activator LpoA | | Authors: | Jean, N.L, Bougault, C, Lodge, A, Derouaux, A, Callens, G, Egan, A, Lewis, R.J, Vollmer, W, Simorre, J. | | Deposit date: | 2013-11-26 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Elongated Structure of the Outer-Membrane Activator of Peptidoglycan Synthesis LpoA: Implications for PBP1A Stimulation.
Structure, 22, 2014
|
|
3LPB
 
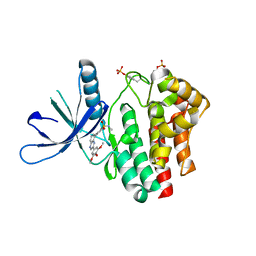 | | Crystal structure of Jak2 complexed with a potent 2,8-diaryl-quinoxaline inhibitor | | Descriptor: | N-methyl-4-[3-(3,4,5-trimethoxyphenyl)quinoxalin-5-yl]benzenesulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Tavares, G.A, Pissot-Soldermann, C, Gerspacher, M, Furet, P, Kroemer, M. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and SAR of potent, orally available 2,8-diaryl-quinoxalines as a new class of JAK2 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3UGC
 
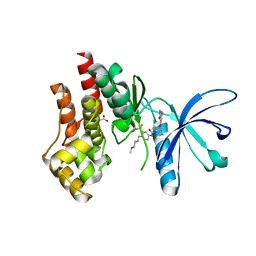 | | Structural basis of Jak2 inhibition by the type II inhibtor NVP-BBT594 | | Descriptor: | 5-{[6-(acetylamino)pyrimidin-4-yl]oxy}-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-2,3-dihydro-1H-indole-1-carboxamide, MALONATE ION, Tyrosine-protein kinase JAK2 | | Authors: | Scheufler, C, Tavares, G.A, Manley, P.W, Pissot-Soldermann, C, Kroemer, M. | | Deposit date: | 2011-11-02 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Modulation of activation-loop phosphorylation by JAK inhibitors is binding mode dependent.
Cancer Discov, 2, 2012
|
|
6TE6
 
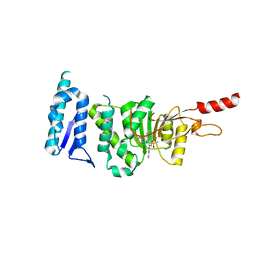 | | Crystal structure of Dot1L in complex with an inhibitor (compound 3). | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, ~{N}1-[(~{S})-(3-chlorophenyl)-pyridin-2-yl-methyl]-4-methylsulfonyl-~{N}2-pyrimidin-2-yl-benzene-1,2-diamine | | Authors: | Scheufler, C, Stauffer, F, Be, C, Moebitz, H. | | Deposit date: | 2019-11-11 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | New Potent DOT1L Inhibitors forin VivoEvaluation in Mouse.
Acs Med.Chem.Lett., 10, 2019
|
|
6TEL
 
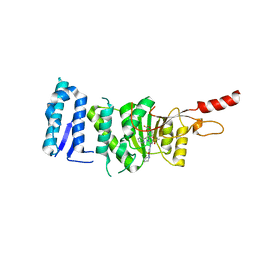 | | Crystal structure of Dot1L in complex with an inhibitor (compound 10). | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, POTASSIUM ION, ... | | Authors: | Scheufler, C, Stauffer, F, Be, C, Moebitz, H. | | Deposit date: | 2019-11-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | New Potent DOT1L Inhibitors forin VivoEvaluation in Mouse.
Acs Med.Chem.Lett., 10, 2019
|
|
6TEN
 
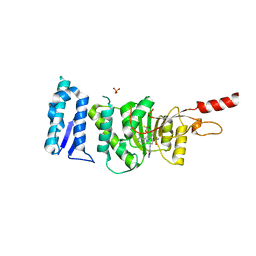 | | Crystal structure of Dot1L in complex with an inhibitor (compound 11). | | Descriptor: | 3-[(4-azanyl-6-methoxy-1,3,5-triazin-2-yl)amino]-4-[[(~{S})-[2,2-bis(fluoranyl)-1,3-benzodioxol-4-yl]-(3-chloranylpyridin-2-yl)methyl]amino]benzenesulfonamide, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Scheufler, C, Stauffer, F, Be, C, Moebitz, H. | | Deposit date: | 2019-11-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | New Potent DOT1L Inhibitors forin VivoEvaluation in Mouse.
Acs Med.Chem.Lett., 10, 2019
|
|
5NWZ
 
 | |
5NUD
 
 | |
5DTR
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD5 [N-(2,6-dichlorophenyl)-4-methoxy-N-methylquinolin-6-amine] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N-(2,6-dichlorophenyl)-4-methoxy-N-methylquinolin-6-amine | | Authors: | Scheufler, C, Be, C, Moebitz, H, Stauffer, F. | | Deposit date: | 2015-09-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Optimization of a Fragment-Based Screening Hit toward Potent DOT1L Inhibitors Interacting in an Induced Binding Pocket.
Acs Med.Chem.Lett., 7, 2016
|
|
5DTQ
 
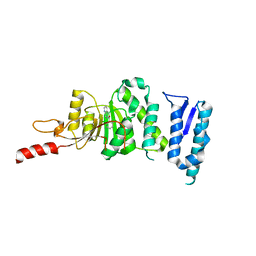 | | Crystal structure of Dot1L in complex with inhibitor CPD3 [(2,6-dichlorophenyl)(quinolin-6-yl)methanone] | | Descriptor: | (2,6-dichlorophenyl)(quinolin-6-yl)methanone, Histone-lysine N-methyltransferase, H3 lysine-79 specific | | Authors: | Scheufler, C, Be, C, Moebitz, H, Stauffer, F. | | Deposit date: | 2015-09-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Optimization of a Fragment-Based Screening Hit toward Potent DOT1L Inhibitors Interacting in an Induced Binding Pocket.
Acs Med.Chem.Lett., 7, 2016
|
|
5DTM
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD1 [4-(2,6-dichlorobenzoyl)-N-methyl-1H-pyrrole-2-carboxamide] | | Descriptor: | 4-(2,6-dichlorobenzoyl)-N-methyl-1H-pyrrole-2-carboxamide, Histone-lysine N-methyltransferase, H3 lysine-79 specific | | Authors: | Scheufler, C, Be, C, Moebitz, H, Stauffer, F. | | Deposit date: | 2015-09-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of a Fragment-Based Screening Hit toward Potent DOT1L Inhibitors Interacting in an Induced Binding Pocket.
Acs Med.Chem.Lett., 7, 2016
|
|
3TXO
 
 | | PKC eta kinase in complex with a naphthyridine | | Descriptor: | 2-methyl-N~1~-[3-(pyridin-4-yl)-2,6-naphthyridin-1-yl]propane-1,2-diamine, Protein kinase C eta type | | Authors: | Stark, W, Rummel, G, Cowan-Jacob, S.W. | | Deposit date: | 2011-09-23 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2,6-Naphthyridines as potent and selective inhibitors of the novel protein kinase C isozymes.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2MTZ
 
 | | Haddock model of Bacillus subtilis L,D-transpeptidase in complex with a peptidoglycan hexamuropeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, Putative L,D-transpeptidase YkuD, intact bacterial peptidoglycan | | Authors: | Schanda, P, Triboulet, S, Laguri, C, Bougault, C, Ayala, I, Callon, M, Arthur, M, Simorre, J. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic model of a cell-wall cross-linking enzyme in complex with an intact bacterial peptidoglycan.
J.Am.Chem.Soc., 136, 2014
|
|
