1NPC
 
 | |
3TXO
 
 | | PKC eta kinase in complex with a naphthyridine | | Descriptor: | 2-methyl-N~1~-[3-(pyridin-4-yl)-2,6-naphthyridin-1-yl]propane-1,2-diamine, Protein kinase C eta type | | Authors: | Stark, W, Rummel, G, Cowan-Jacob, S.W. | | Deposit date: | 2011-09-23 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2,6-Naphthyridines as potent and selective inhibitors of the novel protein kinase C isozymes.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3IW4
 
 | | Crystal structure of PKC alpha in complex with NVP-AEB071 | | Descriptor: | 3-(1H-indol-3-yl)-4-[2-(4-methylpiperazin-1-yl)quinazolin-4-yl]-1H-pyrrole-2,5-dione, Protein kinase C alpha type | | Authors: | Stark, W, Rummel, G, Strauss, A, Cowan-Jacob, S.W. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 3-(1H-indol-3-yl)-4-[2-(4-methylpiperazin-1-yl)quinazolin-4-yl]pyrrole-2,5-dione (AEB071), a potent and selective inhibitor of protein kinase C isotypes
J.Med.Chem., 52, 2009
|
|
1UOM
 
 | | The Structure of Estrogen Receptor in Complex with a Selective and Potent Tetrahydroisochiolin Ligand. | | Descriptor: | 2-PHENYL-1-[4-(2-PIPERIDIN-1-YL-ETHOXY)-PHENYL]-1,2,3,4-TETRAHYDRO-ISOQUINOLIN-6-OL, ESTROGEN RECEPTOR | | Authors: | Stark, W, Bischoff, S.F, Buhl, T, Fournier, B, Halleux, C, Kallen, J, Keller, H, Renaud, J. | | Deposit date: | 2003-04-11 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Estrogen Receptor Modulators: Identification and Structure-Activity Relationships of Potent Eralpha-Selective Tetrahydroisoquinoline Ligands
J.Med.Chem., 46, 2003
|
|
2JED
 
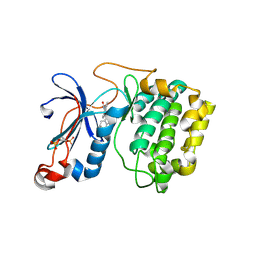 | | The crystal structure of the kinase domain of the protein kinase C theta in complex with NVP-XAA228 at 2.32A resolution. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(8-DIMETHYLAMINOMETHYL-6,7,8,9-TETRAHYDRO-PYRIDO[1,2-A]INDOL-10-YL)-4-(1-METHYL-1H-INDOL-3-YL)-PYRROLE-2,5-DIONE, PROTEIN KINASE C THETA | | Authors: | Stark, W, Bitsch, F, Berner, A, Buelens, F, Graff, P, Depersin, H, Geiser, M, Knecht, R, Rahuel, J, Rummel, G, Schlaeppi, J.M, Schmitz, R, Strauss, A, Wagner, J. | | Deposit date: | 2007-01-16 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The Crystal Structure of the Kinase Domain of the Protein Kinase C Theta in Complex with Nvp-Xaa228
To be Published
|
|
5MVZ
 
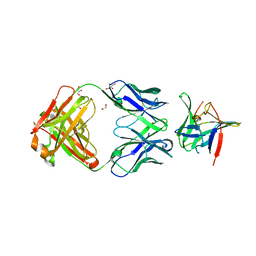 | | Fab 4AB007 bound to Interleukin-1-beta | | Descriptor: | Fab 4AB007 H-chain, Fab 4AB007 L-chain, GLYCEROL, ... | | Authors: | Stark, W, Seibert, V. | | Deposit date: | 2017-01-17 | | Release date: | 2017-02-15 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of a novel, fully Human Fab binding to Interleukin 1-beta.
To Be Published
|
|
5EW3
 
 | |
1BJO
 
 | |
6P93
 
 | | Human APE1 K98A AP-endonuclease product complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Whitaker, A.W, Stark, W.J, Freudenthal, B.D. | | Deposit date: | 2019-06-09 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functions of the major abasic endonuclease (APE1) in cell viability and genotoxin resistance.
Mutagenesis, 35, 2020
|
|
6P94
 
 | | Human APE1 C65A AP-endonuclease product complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Whitaker, A.W, Stark, W.J, Freudenthal, B.D. | | Deposit date: | 2019-06-09 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Functions of the major abasic endonuclease (APE1) in cell viability and genotoxin resistance.
Mutagenesis, 35, 2020
|
|
1J0X
 
 | | Crystal structure of the rabbit muscle glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Cowan-Jacob, S.W, Kaufmann, M, Anselmo, A.N, Stark, W, Grutter, M.G. | | Deposit date: | 2002-11-25 | | Release date: | 2003-12-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of rabbit-muscle glyceraldehyde-3-phosphate dehydrogenase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3PKZ
 
 | | Structural basis for catalytic activation of a serine recombinase | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Recombinase Sin, ... | | Authors: | Keenholtz, R.A, Boocock, M.R, Rowland, S.J, Stark, W.M, Rice, P.A. | | Deposit date: | 2010-11-12 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for catalytic activation of a serine recombinase.
Structure, 19, 2011
|
|
3O77
 
 | | The structure of Ca2+ Sensor (Case-16) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Myosin light chain kinase, ... | | Authors: | Leder, L, Stark, W, Freuler, F, Marsh, M, Meyerhofer, M, Stettler, T, Mayr, L.M, Britanova, O.V, Strukova, L.A, Chudakov, D.M. | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of Ca2+ sensor Case16 reveals the mechanism of reaction to low Ca2+ concentrations
Sensors (Basel), 10, 2010
|
|
3O78
 
 | | The structure of Ca2+ Sensor (Case-12) | | Descriptor: | CALCIUM ION, Myosin light chain kinase, smooth muscle,Green fluorescent protein,Green fluorescent protein,Calmodulin-1 | | Authors: | Leder, L, Stark, W, Freuler, F, Marsh, M, Meyerhofer, M, Stettler, T, Mayr, L.M, Britanova, O.V, Strukova, L.A, Chudakov, D.M. | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of Ca2+ sensor Case16 reveals the mechanism of reaction to low Ca2+ concentrations
Sensors (Basel), 10, 2010
|
|
4C3F
 
 | |
