8Q1T
 
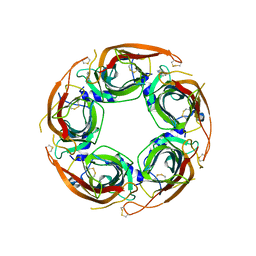 | | X-ray structure of acetylcholine binding protein (AChBP) in complex with IOTA739 | | Descriptor: | 1,10-PHENANTHROLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Cederfelt, D, Lund, B.A, Boronat, P, Hennig, S, Dobritzsch, D, Danielson, U.H. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Elucidating the regulation of ligand gated ion channels via biophysical studies of ligand-induced conformational dynamics of acetylcholine binding proteins
To Be Published
|
|
6YUH
 
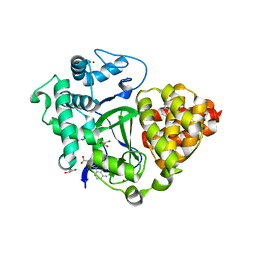 | | Crystal structure of SMYD3 with diperodon R enantiomer bound to allosteric site | | Descriptor: | Diperodon, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Cederfelt, D, Talibov, V.O, Dobritzsch, D, Danielson, U.H. | | Deposit date: | 2020-04-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of an Allosteric Ligand Binding Site in SMYD3 Lysine Methyltransferase.
Chembiochem, 22, 2021
|
|
5CYI
 
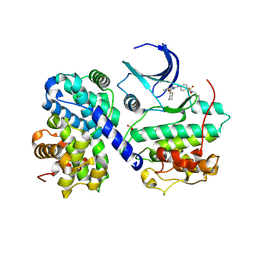 | | CDK2/Cyclin A covalent complex with 6-(cyclohexylmethoxy)-N-(4-(vinylsulfonyl)phenyl)-9H-purin-2-amine (NU6300) | | Descriptor: | 6-(cyclohexylmethoxy)-N-[4-(ethylsulfonyl)phenyl]-9H-purin-2-amine, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Anscombe, E, Meschini, E, Vidal, R.M, Martin, M.P, Staunton, D, Geitmann, M, Danielson, U.H, Stanley, W.A, Wang, L.Z, Reuillon, T, Golding, B.T, Cano, C, Newell, D.R, Noble, M.E.M, Wedge, S.R, Endicott, J.A, Griffin, R.J. | | Deposit date: | 2015-07-30 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and Characterization of an Irreversible Inhibitor of CDK2.
Chem.Biol., 22, 2015
|
|
7NDV
 
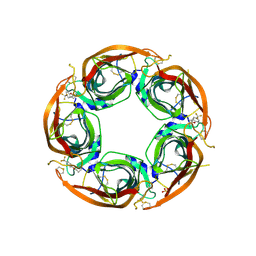 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL001888. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[4-(trifluoromethyl)phenoxy]piperidine, Acetylcholine-binding protein, ... | | Authors: | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | Deposit date: | 2021-02-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of fragments inducing conformational effects in dynamic proteins using a second-harmonic generation biosensor
RSC Advances, 11, 2021
|
|
7NDP
 
 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL001856. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-bromanylspiro[3~{H}-chromene-2,4'-piperidine]-4-one, ... | | Authors: | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | Deposit date: | 2021-02-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of fragments inducing conformational effects in dynamic proteins using a second-harmonic generation biosensor.
Rsc Adv, 11, 2021
|
|
8P1F
 
 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL001909. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-azanyl-1-phenyl-piperidine-4-carboxylic acid, Acetylcholine-binding protein | | Authors: | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elucidating the regulation of ligand gated ion channels via biophysical studies of ligand-induced conformational dynamics of acetylcholine binding proteins
To Be Published
|
|
8P22
 
 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with IOTA376. | | Descriptor: | 2-[(2~{R})-1-ethylimidazolidin-2-yl]-6-pyridin-2-yl-pyridine, Acetylcholine-binding protein, GLYCEROL, ... | | Authors: | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | Deposit date: | 2023-05-14 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Elucidating the regulation of ligand gated ion channels via biophysical studies of ligand-induced conformational dynamics of acetylcholine binding proteins
To Be Published
|
|
8P1E
 
 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL001613. | | Descriptor: | 1-[4-(trifluoromethyl)pyridin-2-yl]piperazine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elucidating the regulation of ligand gated ion channels via biophysical studies of ligand-induced conformational dynamics of acetylcholine binding proteins
To Be Published
|
|
8P11
 
 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL003044. | | Descriptor: | 4-(4-chlorophenyl)piperidin-4-ol, Acetylcholine-binding protein, CHLORIDE ION, ... | | Authors: | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Elucidating the regulation of ligand gated ion channels via biophysical studies of ligand-induced conformational dynamics of acetylcholine binding proteins
To Be Published
|
|
5AFH
 
 | | alpha7-AChBP in complex with lobeline | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN, NEURONAL ACETYLCHOLINE RECEPTOR SUBUNIT ALPHA-7, ... | | Authors: | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AFL
 
 | | alpha7-AChBP in complex with lobeline and fragment 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN, NEURONAL ACETYLCHOLINE RECEPTOR SUBUNIT ALPHA-7, ... | | Authors: | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AFK
 
 | | alpha7-AChBP in complex with lobeline and fragment 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN, NEURONAL ACETYLCHOLINE RECEPTOR SUBUNIT ALPHA-7, ... | | Authors: | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AFN
 
 | | alpha7-AChBP in complex with lobeline and fragment 5 | | Descriptor: | (4R)-4-(2-phenylethyl)pyrrolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN, ... | | Authors: | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AFM
 
 | | alpha7-AChBP in complex with lobeline and fragment 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,5-dibromo-N-(3-hydroxypropyl)-1H-pyrrole-2-carboxamide, ACETYLCHOLINE-BINDING PROTEIN, ... | | Authors: | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AFJ
 
 | | alpha7-AChBP in complex with lobeline and fragment 1 | | Descriptor: | (3S)-6-(4-bromophenyl)-3-hydroxy-1,3-dimethyl-2,3-dihydropyridin-4(1H)-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN, ... | | Authors: | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7BJ1
 
 | | Crystal structure of SMYD3 with diperodon S enantiomer bound to allosteric site | | Descriptor: | ACETATE ION, Diperodon (S-enantiomer), GLYCEROL, ... | | Authors: | Talibov, V.O, Cederfelt, D, Dobritzsch, D, Danielson, U.H. | | Deposit date: | 2021-01-13 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Discovery of an Allosteric Ligand Binding Site in SMYD3 Lysine Methyltransferase
Chembiochem, 22, 2021
|
|
8BBQ
 
 | | Determination of the structure of active tyrosinase from bacterium Verrucomicrobium spinosum | | Descriptor: | COPPER (II) ION, Core tyrosinase, GLYCEROL, ... | | Authors: | Fekry, M, Dave, K, Badgujar, D, Aurelius, O, Hamnevik, E, Dobritzsch, D, Danielson, H. | | Deposit date: | 2022-10-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The Crystal Structure of Tyrosinase from Verrucomicrobium spinosum Reveals It to Be an Atypical Bacterial Tyrosinase.
Biomolecules, 13, 2023
|
|
8BBR
 
 | | Determination of the structure of active tyrosinase from bacterium Verrucomicrobium spinosum | | Descriptor: | COPPER (II) ION, Core tyrosinase, SULFATE ION | | Authors: | Fekry, M, Dave, K, Badgujar, D, Aurelius, O, Hamnevik, E, Dobritzsch, D, Danielson, H. | | Deposit date: | 2022-10-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Crystal Structure of Tyrosinase from Verrucomicrobium spinosum Reveals It to Be an Atypical Bacterial Tyrosinase.
Biomolecules, 13, 2023
|
|
3I5K
 
 | | Crystal structure of the NS5B polymerase from Hepatitis C Virus (HCV) strain JFH1 | | Descriptor: | PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Simister, P.C, Schmitt, M, Lohmann, V, Bressanelli, S. | | Deposit date: | 2009-07-05 | | Release date: | 2009-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of hepatitis C virus strain JFH1 polymerase
J.Virol., 83, 2009
|
|
6S35
 
 | | LSD1/CoREST1 complex with macrocyclic peptide inhibitor | | Descriptor: | ALA-ARG-(D)LYS-MET-GLN-GLU-ALA-ARG-LYS-SER-THR, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, ... | | Authors: | Talibov, V.O, Dobritzsch, D. | | Deposit date: | 2019-06-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Macrocyclic Peptides Uncover a Novel Binding Mode for Reversible Inhibitors of LSD1.
Acs Omega, 5, 2020
|
|
2YME
 
 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with granisetron | | Descriptor: | 1-methyl-N-[(1R,5S)-9-methyl-9-azabicyclo[3.3.1]nonan-3-yl]indazole-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Ligand Recognition in 5-Ht3 Receptors.
Embo Rep., 14, 2013
|
|
2YMD
 
 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with serotonin (5-hydroxytryptamine) | | Descriptor: | GLYCEROL, PHOSPHATE ION, SEROTONIN, ... | | Authors: | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis of Ligand Recognition in 5-Ht(3) Receptors.
Embo Rep., 14, 2013
|
|
7QBB
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 18 | | Descriptor: | 3C-like proteinase nsp5, 7-isoquinolin-4-yl-2-phenyl-5,7-diazaspiro[3.4]octane-6,8-dione, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-11-18 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7O46
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 17 | | Descriptor: | 2-cyclobutyl-7-isoquinolin-4-yl-5,7-diazaspiro[3.4]octane-6,8-dione, 3C-like proteinase nsp5 | | Authors: | Talibov, V.O. | | Deposit date: | 2021-04-05 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
8PT4
 
 | | beta-Ureidopropionase tetramer | | Descriptor: | Beta-ureidopropionase | | Authors: | Cederfelt, D, Dobritzsch, D. | | Deposit date: | 2023-07-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | The Allosteric Regulation of Beta-Ureidopropionase Depends on Fine-Tuned Stability of Active-Site Loops and Subunit Interfaces.
Biomolecules, 13, 2023
|
|
