8GO7
 
 | | Fungal immunomodulatory protein FIP-nha N5+39A | | Descriptor: | Fungal immunomodulatory protein FIP-nha | | Authors: | Liu, Y, Bastiaan-Net, S, Hoppenbrouwers, T, Li, Z, Wichers, H.J. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Glycosylation Contributes to Thermostability and Proteolytic Resistance of rFIP-nha ( Nectria haematococca ).
Molecules, 28, 2023
|
|
8GO5
 
 | |
4QNU
 
 | | Crystal structure of CmoB bound with Cx-SAM in P21212 | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, PHOSPHATE ION, tRNA (mo5U34)-methyltransferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-06-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Determinants of the CmoB carboxymethyl transferase utilized for selective tRNA wobble modification.
Nucleic Acids Res., 43, 2015
|
|
7LD1
 
 | |
7LCN
 
 | |
5WVK
 
 | | Yeast proteasome-ADP-AlFx | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Ding, Z, Cong, Y. | | Deposit date: | 2016-12-25 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx
Cell Res., 27, 2017
|
|
7WG6
 
 | | Neutral Omicron Spike Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Cui, Z, Wang, X. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-18 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
8HBC
 
 | | Crystal structure of the CysR-CTLD3 fragment of human DEC205 | | Descriptor: | Lymphocyte antigen 75 | | Authors: | Kong, D, Yu, B, Hu, Z, Cheng, C, Cao, L, He, Y. | | Deposit date: | 2022-10-28 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Interaction of human dendritic cell receptor DEC205/CD205 with keratins.
J.Biol.Chem., 300, 2024
|
|
4RYP
 
 | |
7V9L
 
 | | Cryo-EM structure of the SV1-Gs complex. | | Descriptor: | GHRH receptor splice variant 1,GHRH receptor splice variant 1,GHRH receptor splice variant 1,SV1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cong, Z.T, Zhou, F.L, Zhang, C, Zou, X.Y, Zhang, H.B, Wang, Y.Z, Zhou, Q.T, Cai, X.Q, Liu, Q.F, Li, J, Shao, L.J, Mao, C.Y, Wang, X, Wu, J.H, Xia, T, Zhao, L.H, Jiang, H.L, Zhang, Y, Xu, H.E, Cheng, X, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-08-26 | | Release date: | 2022-04-06 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Constitutive signal bias mediated by the human GHRHR splice variant 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5EOB
 
 | | Crystal structure of CMET in complex with novel inhibitor | | Descriptor: | 6-[bis(fluoranyl)-[6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl]methyl]quinoline, Hepatocyte growth factor receptor | | Authors: | Liu, Q, Chen, T, Xu, Y. | | Deposit date: | 2015-11-10 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of 6-(difluoro(6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl)methyl)quinoline as a highly potent and selective c-Met inhibitor
Eur.J.Med.Chem., 116, 2016
|
|
4RXD
 
 | |
7LZ7
 
 | | Tubulin-RB3_SLD-TTL in complex with compound 5k | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(3,6-dimethyl[1,2]oxazolo[5,4-d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
7LZ8
 
 | | Tubulin-RB3_SLD-TTL in complex with compound 5t | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[2-(ethylamino)pyrido[3,2-d]pyrimidin-4-yl]-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
4RXE
 
 | |
4RXC
 
 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Homorisedronate BPH-6 | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, [1-hydroxy-3-(pyridin-3-yl)propane-1,1-diyl]bis(phosphonic acid) | | Authors: | Cao, R, Liu, Y.-L, Oldfield, E. | | Deposit date: | 2014-12-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Farnesyl diphosphate synthase inhibitors with unique ligand-binding geometries.
ACS Med Chem Lett, 6, 2015
|
|
7LAB
 
 | |
7LAA
 
 | |
2HNK
 
 | | Crystal structure of SAM-dependent O-methyltransferase from pathogenic bacterium Leptospira interrogans | | Descriptor: | DI(HYDROXYETHYL)ETHER, S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent O-methyltransferase, ... | | Authors: | Hou, X, Wei, Z, Gong, W. | | Deposit date: | 2006-07-13 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SAM-dependent O-methyltransferase from pathogenic bacterium Leptospira interrogans.
J.Struct.Biol., 159, 2007
|
|
6KPF
 
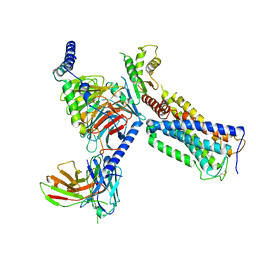 | | Cryo-EM structure of a class A GPCR with G protein complex | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Shen, L, Wang, Y.X, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
7V9M
 
 | | Cryo-EM structure of the GHRH-bound human GHRHR splice variant 1 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Cong, Z.T, Zhou, F.L, Zhang, C, Zou, X.Y, Zhang, H.B, Wang, Y.Z, Zhou, Q.T, Cai, X.Q, Liu, Q.F, Li, J, Shao, L.J, Mao, C.Y, Wang, X, Wu, J.H, Xia, T, Zhao, L.H, Jiang, H.L, Zhang, Y, Xu, H.E, Chen, X, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Constitutive signal bias mediated by the human GHRHR splice variant 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6KPG
 
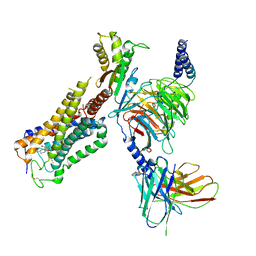 | | Cryo-EM structure of CB1-G protein complex | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hua, T, Li, X.T, Wu, L.J, Makriyannis, A, Wang, Y.X, Shen, L, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
2I6K
 
 | | Crystal structure of human type I IPP isomerase complexed with a substrate analog | | Descriptor: | ACETIC ACID, AMINOETHANOLPYROPHOSPHATE, Isopentenyl-diphosphate delta-isomerase 1, ... | | Authors: | Zhang, C, Wei, Z, Gong, W. | | Deposit date: | 2006-08-29 | | Release date: | 2007-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human IPP isomerase: new insights into the catalytic mechanism
J.Mol.Biol., 366, 2007
|
|
7XUX
 
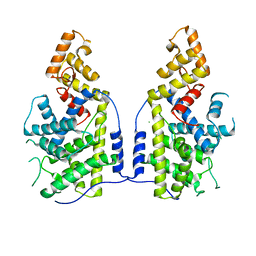 | |
8IND
 
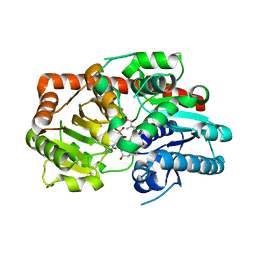 | | Crystal structure of UGT74AN3-UDP-RES | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, ... | | Authors: | Huang, W. | | Deposit date: | 2023-03-09 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
