7SQ8
 
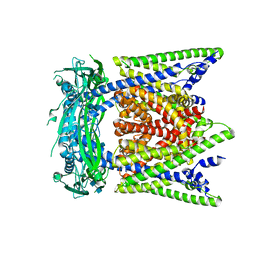 | | Cryo-EM structure of mouse apo TRPML1 channel at 2.598 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, SODIUM ION | | Authors: | Gan, N, Han, Y, Jiang, Y. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.598 Å) | | Cite: | Structural mechanism of allosteric activation of TRPML1 by PI(3,5)P 2 and rapamycin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SQ9
 
 | | Cryo-EM structure of mouse temsirolimus/PI(3,5)P2-bound TRPML1 channel at 2.11 Angstrom resolution | | Descriptor: | (1R,2R,4S)-4-{(2R)-2-[(3S,6R,7E,9R,10R,12R,14S,15E,17E,19E,21S,23S,26R,27R,30S,34aS)-9,27-dihydroxy-10,21-dimethoxy-6,8,12,14,20,26-hexamethyl-1,5,11,28,29-pentaoxo-1,4,5,6,9,10,11,12,13,14,21,22,23,24,25,26,27,28,29,31,32,33,34,34a-tetracosahydro-3H-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontin-3-yl]propyl}-2-methoxycyclohexyl 3-hydroxy-2-(hydroxymethyl)-2-methylpropanoate, (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gan, N, Han, Y, Jiang, Y. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Structural mechanism of allosteric activation of TRPML1 by PI(3,5)P 2 and rapamycin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SQ6
 
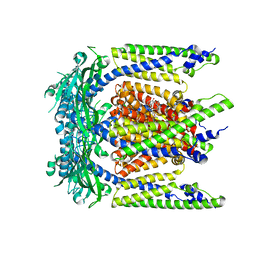 | | Cryo-EM structure of mouse agonist ML-SA1-bound TRPML1 channel at 2.32 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-oxo-2-[(4S)-2,2,4-trimethyl-3,4-dihydroquinolin-1(2H)-yl]ethyl}-1H-isoindole-1,3(2H)-dione, Mucolipin-1, ... | | Authors: | Gan, N, Han, Y, Jiang, Y. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | Structural mechanism of allosteric activation of TRPML1 by PI(3,5)P 2 and rapamycin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4XAK
 
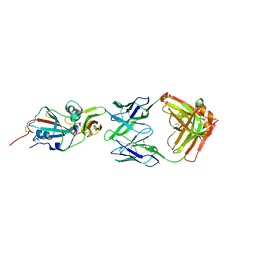 | | Crystal structure of potent neutralizing antibody m336 in complex with MERS Co-V RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Heavy chain of neutralizing antibody m336, ... | | Authors: | Zhou, T, Dimtrov, D.S, Ying, T. | | Deposit date: | 2014-12-15 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody.
Nat Commun, 6, 2015
|
|
8Z5E
 
 | |
8Z5D
 
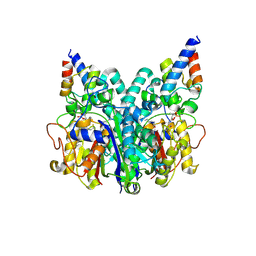 | |
8Z5F
 
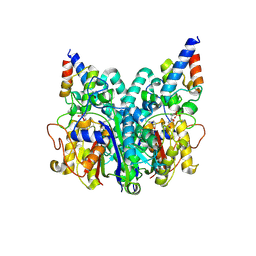 | |
7SC5
 
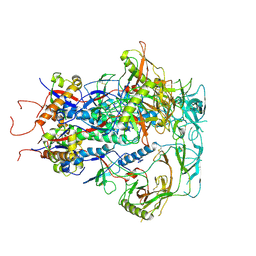 | | Cytoplasmic tail deleted HIV Env trimer in nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Yang, S, Walz, T. | | Deposit date: | 2021-09-27 | | Release date: | 2022-11-09 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Dynamic HIV-1 spike motion creates vulnerability for its membrane-bound tripod to antibody attack.
Nat Commun, 13, 2022
|
|
7SD3
 
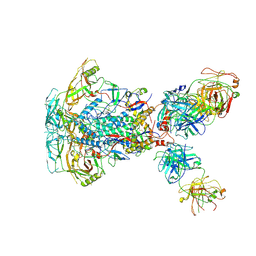 | | Cytoplasmic tail deleted HIV-1 Env bound with three 4E10 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4E10 Fab heavy chain, ... | | Authors: | Yang, S, Walz, T. | | Deposit date: | 2021-09-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Dynamic HIV-1 spike motion creates vulnerability for its membrane-bound tripod to antibody attack.
Nat Commun, 13, 2022
|
|
7BVC
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVF
 
 | | Cryo-EM structure of Mycobacterium tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, CARDIOLIPIN, Ethambutol, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVE
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbC2-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, Ethambutol, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVG
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with di-arabinose. | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVH
 
 | | Crystal structure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, Meromycolate extension acyl carrier protein, ... | | Authors: | Zhao, Y, Zhang, L, Wu, L.J, Wang, Q, Li, J, Besra, G.S, Rao, Z.H. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BYR
 
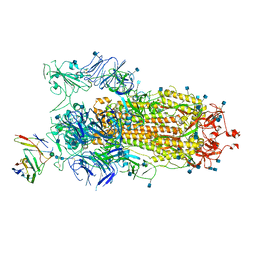 | | BD23-Fab in complex with the S ectodomain trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab23-Fab-Heavy Chain, ... | | Authors: | Zhu, Q, Wang, G, Xiao, J. | | Deposit date: | 2020-04-24 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients' B Cells.
Cell, 182, 2020
|
|
5XHS
 
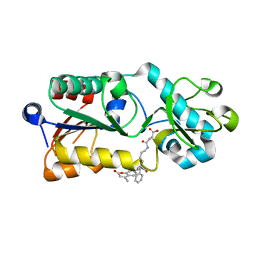 | | Crystal structure of SIRT5 complexed with a fluorogenic small-molecule substrate SuBKA | | Descriptor: | (2S)-2-azanyl-6-[(4-hydroxy-4-oxo-butanoyl)amino]hexanoic acid, 7-AMINO-4-METHYL-CHROMEN-2-ONE, NAD-dependent protein deacylase sirtuin-5, ... | | Authors: | Yu, Y, Li, B, Chen, Q. | | Deposit date: | 2017-04-24 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Interactions between sirtuins and fluorogenic small-molecule substrates offer insights into inhibitor design
Rsc Adv, 7, 2017
|
|
7S0J
 
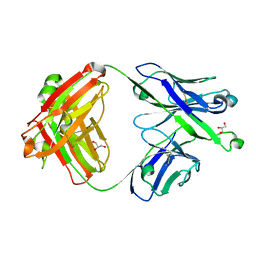 | | Crystal structure of Epstein-Barr virus gH/gL targeting antibody 769B10 | | Descriptor: | 769B10 Fab Heavy chain, 769B10 Fab Light chain, GLYCEROL | | Authors: | Chen, W.-H, Kanekiyo, M, Cohen, J.I, Joyce, M.G. | | Deposit date: | 2021-08-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
7S08
 
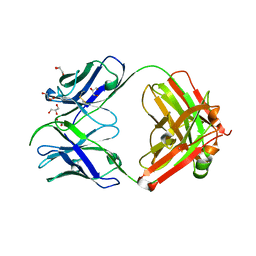 | |
7S1B
 
 | | Crystal structure of Epstein-Barr virus glycoproteins gH/gL/gp42-peptide in complex with human neutralizing antibodies 769C2 and 770F7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769C2 Fab heavy chain, 769C2 Fab light chain, ... | | Authors: | Chen, W.-H, Cohen, J.I, Kanekiyo, M, Joyce, M.G. | | Deposit date: | 2021-09-02 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
7S07
 
 | | Crystal structure of Epstein-Barr virus glycoprotein gH/gL/gp42-peptide in complex with human neutralizing antibodies 769B10 and 769C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769B10 Fab heavy chain, 769B10 Fab light chain, ... | | Authors: | Chen, W.-H, Kanekiyo, M, Cohen, J.I, Joyce, M.G. | | Deposit date: | 2021-08-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
7L3N
 
 | | SARS-CoV 2 Spike Protein bound to LY-CoV555 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV555 Fab heavy chain, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection.
Biorxiv, 2020
|
|
5OPB
 
 | | Structure of CHK1 10-pt. mutant complex with indazole LRRK2 inhibitor | | Descriptor: | (2~{R},6~{S})-2,6-dimethyl-4-[6-[5-(1-methylcyclopropyl)oxy-1~{H}-indazol-3-yl]pyrimidin-4-yl]morpholine, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OQ8
 
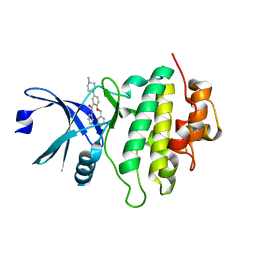 | | Structure of CHK1 12-pt. mutant complex with arylbenzamide LRRK2 inhibitor | | Descriptor: | 5-(4-methylpiperazin-1-yl)-2-phenylmethoxy-~{N}-pyridin-3-yl-benzamide, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
6QHA
 
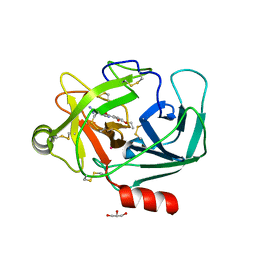 | | Crystal Structure of Human Kallikrein 6 in complex with GSK3205388B | | Descriptor: | GLYCEROL, Kallikrein-6, UNKNOWN ATOM OR ION, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Kallikrein 5 inhibitors identified through structure based drug design in search for a treatment for Netherton Syndrome.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6QFF
 
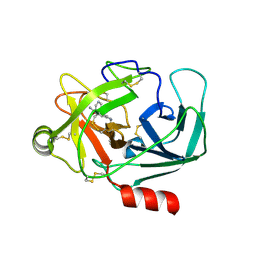 | | Crystal Structure of Human Kallikrein 6 in complex with GSK144 | | Descriptor: | 4-[(5-phenyl-1~{H}-imidazol-2-yl)methylamino]-2-(pyridin-3-ylmethoxy)benzenecarboximidamide, GLYCEROL, Kallikrein-6 | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Evaluation of a crystallographic surrogate for kallikrein 5 in the discovery of novel inhibitors for Netherton syndrome.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
