3PPW
 
 | | Crystal structure of the D1596A mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
7DMC
 
 | | Dipyridamole binds to the N-terminal domain of human Hsp90A | | Descriptor: | 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, CHLORIDE ION, Heat shock protein HSP 90-alpha, ... | | Authors: | Shi, L, Zhou, C, Zhong, Y, Gao, J, Zhou, H, Zhang, N. | | Deposit date: | 2020-12-03 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Dipyridamole interacts with the N-terminal domain of HSP90 and antagonizes the function of the chaperone in multiple cancer cell lines.
Biochem Pharmacol, 207, 2022
|
|
7DWA
 
 | | Structure of a novel beta-mannanase BaMan113A with mannotriose, N236Y mutation | | Descriptor: | Endo-beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Liu, W.T, Liu, W.D, Zheng, Y.Y. | | Deposit date: | 2021-01-15 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Functional and structural investigation of a novel beta-mannanase BaMan113A from Bacillus sp. N16-5.
Int.J.Biol.Macromol., 182, 2021
|
|
7DW8
 
 | | Structure of a novel beta-mannanase BaMan113A with mannobiose, N236Y mutation. | | Descriptor: | Endo-beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Liu, W.T, Liu, W.D, Zheng, Y.Y. | | Deposit date: | 2021-01-15 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and structural investigation of a novel beta-mannanase BaMan113A from Bacillus sp. N16-5.
Int.J.Biol.Macromol., 182, 2021
|
|
7DVJ
 
 | | Structure of beta-mannanase BaMan113A with mannobiose | | Descriptor: | Endo-beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Liu, W.T, Liu, W.D, Zheng, Y.Y. | | Deposit date: | 2021-01-13 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional and structural investigation of a novel beta-mannanase BaMan113A from Bacillus sp. N16-5.
Int.J.Biol.Macromol., 182, 2021
|
|
7DVZ
 
 | |
7DV7
 
 | |
2KVQ
 
 | |
3TG5
 
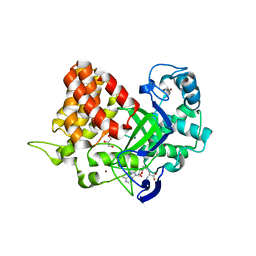 | | Structure of SMYD2 in complex with p53 and SAH | | Descriptor: | Cellular tumor antigen p53, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | Authors: | Zhao, K, Wang, L. | | Deposit date: | 2011-08-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human SMYD2 reveals the basis of p53 tumor suppressor methylation
J.Biol.Chem., 2011
|
|
3TG4
 
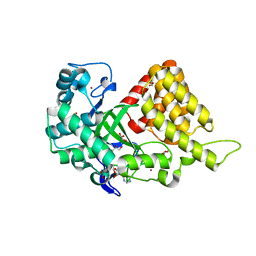 | | Structure of SMYD2 in complex with SAM | | Descriptor: | GLYCEROL, N-lysine methyltransferase SMYD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Zhao, K, Wang, L. | | Deposit date: | 2011-08-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human SMYD2 reveals the basis of p53 tumor suppressor methylation
J.Biol.Chem., 2011
|
|
6J9A
 
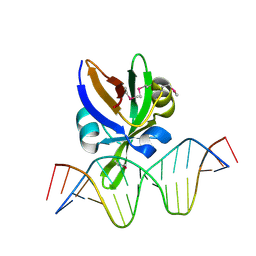 | | Crystal structure of Arabidopsis thaliana VAL1 in complex with FLC DNA fragment | | Descriptor: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*AP*TP*C)-3'), DNA (5'-D(*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*TP*TP*G)-3') | | Authors: | Hu, H, Du, J. | | Deposit date: | 2019-01-22 | | Release date: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.915 Å) | | Cite: | Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis.
Nat.Plants, 5, 2019
|
|
6J9C
 
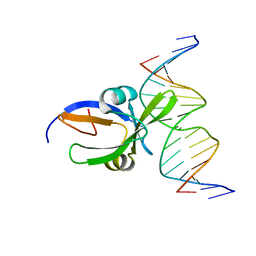 | | Crystal structure of Arabidopsis thaliana transcription factor LEC2-DNA complex | | Descriptor: | B3 domain-containing transcription factor LEC2, DNA (5'-D(*CP*AP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*AP*T)-3'), DNA (5'-D(*GP*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*TP*T)-3') | | Authors: | Hu, H, Du, J. | | Deposit date: | 2019-01-22 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis.
Nat.Plants, 5, 2019
|
|
8JGG
 
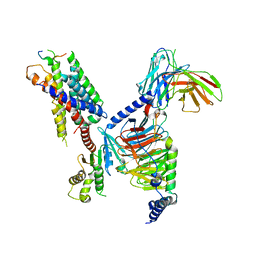 | | CryoEM structure of Gi-coupled MRGPRX1 with peptide agonist BAM8-22 | | Descriptor: | BAM8-22, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Xu, H.E, Ynag, F, Liu, Z.M, Guo, L.L, Zhang, Y.M, Fang, G.X, Tie, L, Zhuang, Y.M, Xue, C.Y. | | Deposit date: | 2023-05-20 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ligand recognition and G protein coupling of the human itch receptor MRGPRX1.
Nat Commun, 14, 2023
|
|
8JGB
 
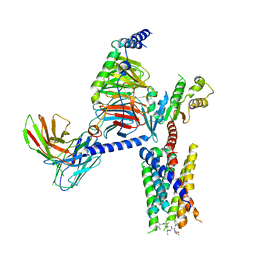 | | CryoEM structure of Gi-coupled MRGPRX1 with peptide agonist CNF-Tx2 | | Descriptor: | Conorfamide-Tx2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Xu, H.E, Yang, F, Liu, Z.M, Guo, L.L, Zhang, Y.M, Fang, G.X, Tie, L, Zhuang, Y.M, Xue, C.Y. | | Deposit date: | 2023-05-20 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Ligand recognition and G protein coupling of the human itch receptor MRGPRX1.
Nat Commun, 14, 2023
|
|
6J9B
 
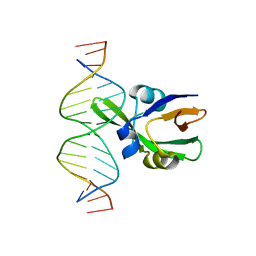 | | Arabidopsis FUS3-DNA complex | | Descriptor: | B3 domain-containing transcription factor FUS3, DNA (5'-D(*AP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*AP*TP*C)-3'), DNA (5'-D(*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*TP*TP*G)-3') | | Authors: | Hu, H, Du, J. | | Deposit date: | 2019-01-22 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis.
Nat.Plants, 5, 2019
|
|
8JGF
 
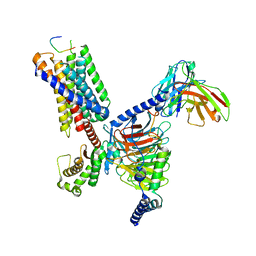 | | CryoEM structure of Gq-coupled MRGPRX1 with peptide agonist BAM8-22 | | Descriptor: | BAM8-22, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Xu, H.E, Yang, F, Liu, Z.M, Guo, L.L, Zhang, Y.M, Fang, G.X, Tie, L, Zhuang, Y.M, Xue, C.Y. | | Deposit date: | 2023-05-20 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Ligand recognition and G protein coupling of the human itch receptor MRGPRX1.
Nat Commun, 14, 2023
|
|
7Y4F
 
 | | bacterial DPP4 | | Descriptor: | Dipeptidyl peptidase IV | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
7Y4G
 
 | | sit-bound btDPP4 | | Descriptor: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, btDPP4 | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
7VS4
 
 | | Crystal structure of PacII_M1M2S-DNA(m6A)-SAH complex | | Descriptor: | DNA (25-mer), S-ADENOSYL-L-HOMOCYSTEINE, Site-specific DNA recognition subunit, ... | | Authors: | Zhu, J, Gao, P. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular insights into DNA recognition and methylation by non-canonical type I restriction-modification systems.
Nat Commun, 13, 2022
|
|
7VRU
 
 | | Crystal structure of PacII_M1M2S-DNA-SAH complex | | Descriptor: | DNA (25-mer), S-ADENOSYL-L-HOMOCYSTEINE, Site-specific DNA recognition subunit, ... | | Authors: | Zhu, J, Gao, P. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into DNA recognition and methylation by non-canonical type I restriction-modification systems.
Nat Commun, 13, 2022
|
|
2UXN
 
 | | Structural Basis of Histone Demethylation by LSD1 Revealed by Suicide Inactivation | | Descriptor: | CHLORIDE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Yang, M, Culhane, J.C, Szewczuk, L.M, Gocke, C.B, Brautigam, C.A, Tomchick, D.R, Machius, M, Cole, P.A, Yu, H. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis of Histone Demethylation by Lsd1 Revealed by Suicide Inactivation.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2UXX
 
 | | Human LSD1 Histone Demethylase-CoREST in complex with an FAD- tranylcypromine adduct | | Descriptor: | CHLORIDE ION, FAD-trans-2-Phenylcyclopropylamine Adduct, GLYCEROL, ... | | Authors: | Yang, M, Culhane, J.C, Machius, M, Cole, P.A, Yu, H. | | Deposit date: | 2007-03-30 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Basis for the Inhibition of the Lsd1 Histone Demethylase by the Antidepressant Trans-2-Phenylcyclopropylamine.
Biochemistry, 46, 2007
|
|
