1I1C
 
 | | NON-FCRN BINDING FC FRAGMENT OF RAT IGG2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IG GAMMA-2A CHAIN C REGION | | Authors: | Martin, W.L, West Jr, A.P, Gan, L, Bjorkman, P.J. | | Deposit date: | 2001-01-31 | | Release date: | 2001-02-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure at 2.8 A of an FcRn/heterodimeric Fc complex: mechanism of pH-dependent binding.
Mol.Cell, 7, 2001
|
|
1I1A
 
 | | CRYSTAL STRUCTURE OF THE NEONATAL FC RECEPTOR COMPLEXED WITH A HETERODIMERIC FC | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Martin, W.L, West Jr, A.P, Gan, L, Bjorkman, P.J. | | Deposit date: | 2001-01-31 | | Release date: | 2001-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure at 2.8 A of an FcRn/heterodimeric Fc complex: mechanism of pH-dependent binding.
Mol.Cell, 7, 2001
|
|
1P7Q
 
 | | Crystal Structure of HLA-A2 Bound to LIR-1, a Host and Viral MHC Receptor | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Willcox, B.E, Thomas, L.M, Bjorkman, P.J. | | Deposit date: | 2003-05-05 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor.
Nat.Immunol., 4, 2003
|
|
7RYV
 
 | |
7RYU
 
 | |
7SC1
 
 | |
7TFN
 
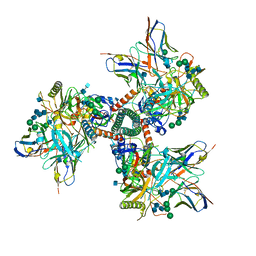 | | Cryo-EM structure of CD4bs antibody Ab1303 in complex with HIV-1 Env trimer BG505 SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-HIV-1 CD4bs antibody Fab Ab1303 - Heavy chain, ... | | Authors: | Yang, Z, Bjorkman, P.J. | | Deposit date: | 2022-01-06 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Neutralizing antibodies induced in immunized macaques recognize the CD4-binding site on an occluded-open HIV-1 envelope trimer.
Nat Commun, 13, 2022
|
|
7TFO
 
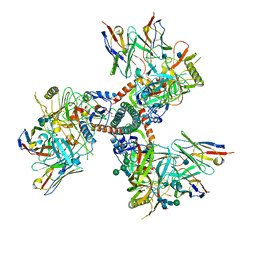 | | Cryo-EM structure of HIV-1 Env trimer BG505 SOSIP.664 in complex with CD4bs antibody Ab1573 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD4 binding site antibody Ab1573 - Fab heavy chain, ... | | Authors: | Yang, Z, Bjorkman, P.J. | | Deposit date: | 2022-01-06 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Neutralizing antibodies induced in immunized macaques recognize the CD4-binding site on an occluded-open HIV-1 envelope trimer.
Nat Commun, 13, 2022
|
|
7U0C
 
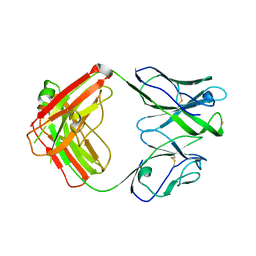 | | Crystal structure of broadly neutralizing antibody HEPC3.4 | | Descriptor: | HEPC3.4 Fab Heavy Chain, HEPC3.4 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J. | | Deposit date: | 2022-02-17 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Computational identification of HCV neutralizing antibodies with a common HCDR3 disulfide bond motif in the antibody repertoires of infected individuals.
Nat Commun, 13, 2022
|
|
7U0B
 
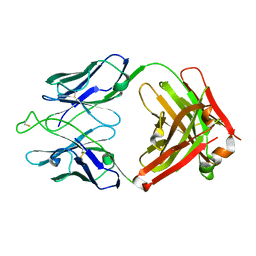 | | Crystal structure of broadly neutralizing antibody HEPC3.1 | | Descriptor: | HEPC3.1 Fab Heavy Chain, HEPC3.1 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J. | | Deposit date: | 2022-02-17 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Computational identification of HCV neutralizing antibodies with a common HCDR3 disulfide bond motif in the antibody repertoires of infected individuals.
Nat Commun, 13, 2022
|
|
1XED
 
 | |
1Z8L
 
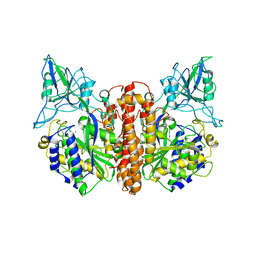 | | Crystal structure of prostate-specific membrane antigen, a tumor marker and peptidase | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Davis, M.I, Bennett, M.J, Thomas, L.M, Bjorkman, P.J. | | Deposit date: | 2005-03-30 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of prostate-specific membrane antigen, a tumor marker and peptidase
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1Z6O
 
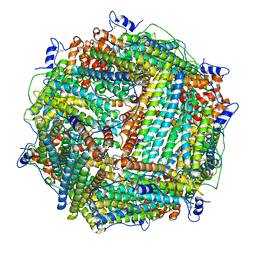 | | Crystal Structure of Trichoplusia ni secreted ferritin | | Descriptor: | CALCIUM ION, FE (III) ION, Ferritin heavy chain, ... | | Authors: | Hamburger, A.E, West Jr, A.P, Hamburger, Z.A, Hamburger, P, Bjorkman, P.J. | | Deposit date: | 2005-03-22 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of a secreted insect ferritin reveals a symmetrical arrangement of heavy and light chains.
J.Mol.Biol., 349, 2005
|
|
1W9R
 
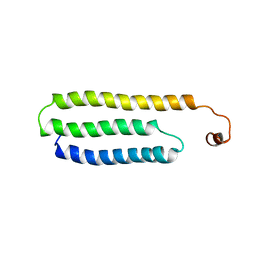 | | Solution Structure of Choline Binding Protein A, Domain R2, the Major Adhesin of Streptococcus pneumoniae | | Descriptor: | CHOLINE BINDING PROTEIN A | | Authors: | Luo, R, Mann, B, Lewis, W.S, Rowe, A, Heath, R, Stewart, M.L, Hamburger, A.E, Bjorkman, P.J, Sivakolundu, S, Lacy, E.R, Tuomanen, E, Kriwacki, R.W. | | Deposit date: | 2004-10-15 | | Release date: | 2005-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Choline Binding Protein A, the Major Adhesin of Streptococcus Pneumoniae
Embo J., 24, 2005
|
|
1ZS8
 
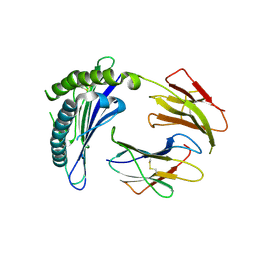 | | Crystal Structure of the Murine MHC Class Ib Molecule M10.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, histocompatibility 2, ... | | Authors: | Olson, R, Huey-Tubman, K.E, Dulac, C, Bjorkman, P.J. | | Deposit date: | 2005-07-22 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a pheromone receptor-associated MHC molecule with an open and empty groove.
Plos Biol., 3, 2005
|
|
2GIY
 
 | |
2GSG
 
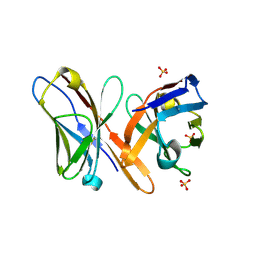 | | Crystal structure of the Fv fragment of a monoclonal antibody specific for poly-glutamine | | Descriptor: | SULFATE ION, monoclonal antibody heavy chain, monoclonal antibody light chain | | Authors: | Li, P, Huey-Tubman, K.E, West Jr, A.P, Bennett, M.J, Bjorkman, P.J. | | Deposit date: | 2006-04-26 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of a polyQ-anti-polyQ complex reveals binding according to a linear lattice model.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2GW5
 
 | | Crystal Structure of LIR-2 (ILT4) at 1.8 : differences from LIR-1 (ILT2) in regions implicated in the binding of the Cytomegalovirus class I MHC homolog UL18 | | Descriptor: | ISOPROPYL ALCOHOL, Leukocyte immunoglobulin-like receptor subfamily B member 2 precursor | | Authors: | Willcox, B.E, Thomas, L.M, Chapman, T.L, Heikema, A.P, West, A.P, Bjorkman, P.J. | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of LIR-2 (ILT4) at 1.8 A: differences from LIR-1 (ILT2) in regions implicated in the binding of the Human Cytomegalovirus class I MHC homolog UL18.
Bmc Struct.Biol., 2, 2002
|
|
2GJ7
 
 | | Crystal Structure of a gE-gI/Fc complex | | Descriptor: | Glycoprotein E, Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sprague, E.R, Wang, C, Baker, D, Bjorkman, P.J. | | Deposit date: | 2006-03-30 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Crystal Structure of the HSV-1 Fc Receptor Bound to Fc Reveals a Mechanism for Antibody Bipolar Bridging.
Plos Biol., 4, 2006
|
|
4JKP
 
 | |
4JDT
 
 | | Crystal structure of chimeric germ-line precursor of NIH45-46 Fab in complex with gp120 of 93TH057 HIV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Scharf, L, Diskin, R, Bjorkman, P.J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JDV
 
 | | Crystal structure of germ-line precursor of NIH45-46 Fab | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Diskin, R, Scharf, L, Bjorkman, P.J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3RPI
 
 | | Crystal Structure of Fab from 3BNC60, Highly Potent anti-HIV Antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain from highly potent anti-HIV neutralizing antibody, Light chain from highly potent anti-HIV neutralizing antibody | | Authors: | Diskin, R, Bjorkman, P.J. | | Deposit date: | 2011-04-26 | | Release date: | 2011-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding.
Science, 333, 2011
|
|
3S3Z
 
 | |
3S3Y
 
 | |
