2G47
 
 | |
3OFI
 
 | | Crystal structure of human insulin-degrading enzyme in complex with ubiquitin | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Insulin-degrading enzyme, Ubiquitin, ... | | Authors: | Kalas, V, Ralat, L.A, Tang, W.-J. | | Deposit date: | 2010-08-15 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Ubiquitin is a novel substrate for human insulin-degrading enzyme.
J.Mol.Biol., 406, 2011
|
|
5COY
 
 | | Crystal structure of CC chemokine 5 (CCL5) | | Descriptor: | C-C motif chemokine 5, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Liang, W.G, Tang, W. | | Deposit date: | 2015-07-20 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.443 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CMD
 
 | |
4IFH
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM44619 | | Descriptor: | Insulin-degrading enzyme, N-({1-[(2R)-4-(hydroxyamino)-1-(naphthalen-2-yl)-4-oxobutan-2-yl]-1H-1,2,3-triazol-4-yl}methyl)-4-methylbenzamide, ZINC ION | | Authors: | Liang, W.G, Guo, Q, Deprez, R, Deprez, B, Tang, W. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.286 Å) | | Cite: | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
5DNF
 
 | | Crystal structure of CC chemokine 5 (CCL5) oligomer in complex with heparin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, C-C motif chemokine 5, ... | | Authors: | Liang, W.G, Tang, W. | | Deposit date: | 2015-09-10 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D65
 
 | | X-RAY STRUCTURE OF MACROPHAGE INFLAMMATORY PROTEIN-1 ALPHA (CCL3) WITH HEPARIN COMPLEX | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, C-C motif chemokine 3, CHLORIDE ION, ... | | Authors: | Liang, W.G, Hwang, D.Y, Zulueta, M.M, Hung, S.C, Tang, W. | | Deposit date: | 2015-08-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4Z6G
 
 | | Structure of NT domain | | Descriptor: | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5, PHOSPHATE ION | | Authors: | Yang, F, Zhang, Y. | | Deposit date: | 2015-04-05 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | In vivo epidermal migration requires focal adhesion targeting of ACF7.
Nat Commun, 7, 2016
|
|
4NXO
 
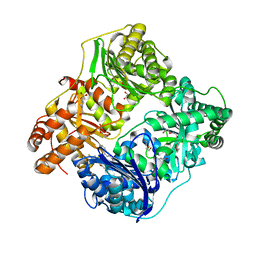 | | Crystal Structure of Insulin Degrading Enzyme in complex with BDM44768 | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liang, W.G, Deprez, R, Deprez, B, Tang, W. | | Deposit date: | 2013-12-09 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
1LVC
 
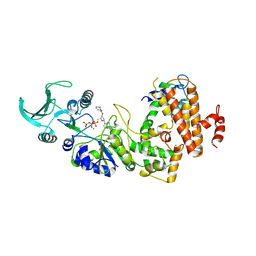 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) in complex with calmodulin and 2' deoxy, 3' anthraniloyl ATP | | Descriptor: | 3'ANTHRANILOYL-2'-DEOXY-ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, YTTERBIUM (III) ION, ... | | Authors: | Shen, Y, Lee, Y.-S, Soelaiman, S, Bergson, P, Lu, D, Chen, A, Beckingham, K, Grabarek, Z, Mrksich, M, Tang, W.-J. | | Deposit date: | 2002-05-28 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Physiological calcium concentrations regulate calmodulin binding and catalysis of adenylyl cyclase exotoxins
Embo J., 21, 2002
|
|
6EDS
 
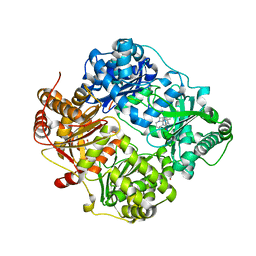 | | Structure of Cysteine-free Human Insulin-Degrading Enzyme in complex with Glucagon and Substrate-selective Macrocyclic Inhibitor 63 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, G.A, Seeliger, M.A, Maianti, J.P, Liu, D.R, Welsh, A.J. | | Deposit date: | 2018-08-10 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.18071723 Å) | | Cite: | Substrate-selective inhibitors that reprogram the activity of insulin-degrading enzyme.
Nat.Chem.Biol., 15, 2019
|
|
8T7I
 
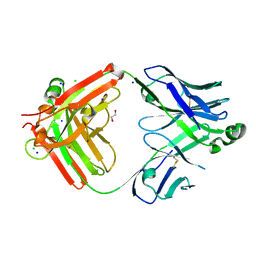 | | Structure of the S1CE variant of Fab F1 (FabS1CE-F1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, S1CE variant of Fab F1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7G
 
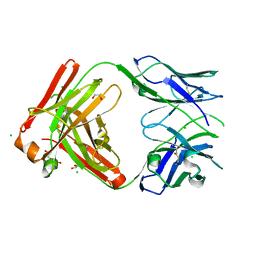 | | Structure of the CK variant of Fab F1 (FabC-F1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CK variant of Fab F1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7F
 
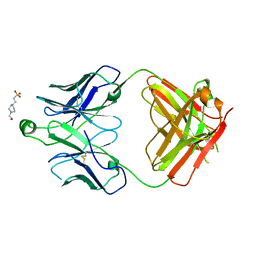 | | Structure of the S1 variant of Fab F1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, S1 variant of Fab F1 heavy chain, S1 variant of Fab F1 light chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
6MQ3
 
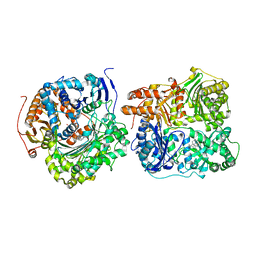 | | Structure of Cysteine-free Human Insulin-Degrading Enzyme in complex with Substrate-selective Macrocycle Inhibitor 63 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Insulin-degrading enzyme, {(8R,9S,10S)-9-(2',3'-dimethyl[1,1'-biphenyl]-4-yl)-6-[(1-methyl-1H-imidazol-2-yl)sulfonyl]-1,6-diazabicyclo[6.2.0]decan-10-yl}methanol | | Authors: | Tan, G.A, Seeliger, M.A, Welsh, A.J, Maianti, J.P, Liu, D.R. | | Deposit date: | 2018-10-09 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.569147 Å) | | Cite: | Substrate-selective inhibitors that reprogram the activity of insulin-degrading enzyme.
Nat.Chem.Biol., 15, 2019
|
|
6AZ2
 
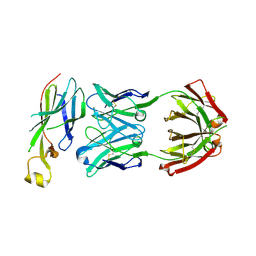 | | Crystal structure of Asf1-Fab 12E complex | | Descriptor: | Fab Heavy Chain, Fab Light Chain, Histone chaperone ASF1 | | Authors: | Bailey, L.J, Kossiakoff, A.A. | | Deposit date: | 2017-09-09 | | Release date: | 2018-01-10 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.477 Å) | | Cite: | Locking the Elbow: Improved Antibody Fab Fragments as Chaperones for Structure Determination.
J. Mol. Biol., 430, 2018
|
|
6AYZ
 
 | | Crystal structure of Asf1-Fab 12E complex | | Descriptor: | Fab Heavy Chain, Fab Light Chain, Histone chaperone ASF1 | | Authors: | Bailey, L.J, Kossiakoff, A.A. | | Deposit date: | 2017-09-08 | | Release date: | 2018-01-17 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Locking the Elbow: Improved Antibody Fab Fragments as Chaperones for Structure Determination.
J. Mol. Biol., 430, 2018
|
|
4LTE
 
 | | Structure of Cysteine-free Human Insulin Degrading Enzyme in Complex with Macrocyclic Inhibitor | | Descriptor: | 2,6-DIAMINO-HEXANOIC ACID AMIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FUMARIC ACID, ... | | Authors: | Foda, Z.H, Seeliger, M.A, Saghatelian, A, Liu, D.R. | | Deposit date: | 2013-07-23 | | Release date: | 2014-05-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Anti-diabetic activity of insulin-degrading enzyme inhibitors mediated by multiple hormones.
Nature, 511, 2014
|
|
