1XSP
 
 | | Crystal Structure of human DNA polymerase lambda in complex with nicked DNA and pyrophosphate | | Descriptor: | 5'-D(*CP*AP*GP*TP*AP*CP*G)-3', 5'-D(*CP*GP*GP*CP*CP*GP*TP*AP*CP*TP*G)-3', 5'-D(P*GP*CP*CP*G)-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Kunkel, T.A, Pedersen, L.C. | | Deposit date: | 2004-10-19 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A closed conformation for the Pol lambda catalytic cycle.
Nat.Struct.Mol.Biol., 12, 2005
|
|
8U0P
 
 | | Synaptic complex of human DNA polymerase Lambda DL variant engaged on a noncomplementary DNA double-strand break | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Kunkel, T.A, Chiruvella, K.K, Ramsden, D.A. | | Deposit date: | 2023-08-29 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA polymerase lambda Loop1 variant yields unexpected gain-of-function capabilities in nonhomologous end-joining.
DNA Repair (Amst), 136, 2024
|
|
8U0O
 
 | | Synaptic complex of human DNA polymerase Lambda DL variant engaged on a DNA double-strand break containing an unpaired 3' primer terminus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*AP*CP*GP*CP*GP*GP*CP*A)-3'), ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Kunkel, T.A, Chiruvella, K.K, Ramsden, D.A. | | Deposit date: | 2023-08-29 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | DNA polymerase lambda Loop1 variant yields unexpected gain-of-function capabilities in nonhomologous end-joining.
DNA Repair (Amst), 136, 2024
|
|
4JWM
 
 | | Ternary complex of D256E mutant of DNA Polymerase Beta | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Perera, L, Ping, L, Shock, D.D, Beard, W.A, Pedersen, L.C, Pedersen, L.G, Wilson, S.H. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Amino Acid Substitution in the Active Site of DNA Polymerase beta Explains the Energy Barrier of the Nucleotidyl Transfer Reaction.
J.Am.Chem.Soc., 135, 2013
|
|
4JWN
 
 | | Ternary complex of D256A mutant of DNA Polymerase Beta | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Perera, L, Ping, L, Shock, D.D, Beard, W.A, Pedersen, L.C, Pedersen, L.G, Wilson, S.H. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Amino Acid Substitution in the Active Site of DNA Polymerase beta Explains the Energy Barrier of the Nucleotidyl Transfer Reaction.
J.Am.Chem.Soc., 135, 2013
|
|
1AQU
 
 | |
3BD9
 
 | | human 3-O-sulfotransferase isoform 5 with bound PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Xu, D, Moon, A.F, Song, D, Liu, J, Pedersen, L.C. | | Deposit date: | 2007-11-14 | | Release date: | 2008-01-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering sulfotransferases to modify heparan sulfate.
Nat.Chem.Biol., 4, 2008
|
|
3C2M
 
 | | Ternary complex of DNA POLYMERASE BETA with a G:dAPCPP mismatch in the active site | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]adenosine, DNA (5'-D(*DCP*DCP*DGP*DAP*DCP*DGP*DGP*DCP*DGP*DCP*DAP*DTP*DCP*DAP*DGP*DC)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of DNA polymerase beta with active-site mismatches suggest a transient abasic site intermediate during misincorporation.
Mol.Cell, 30, 2008
|
|
3C2K
 
 | | DNA POLYMERASE BETA with a gapped DNA substrate and DUMPNPP with Manganese in the active site | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*DCP*DCP*DGP*DAP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DCP*DAP*DGP*DC)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of DNA polymerase beta with active-site mismatches suggest a transient abasic site intermediate during misincorporation.
Mol.Cell, 30, 2008
|
|
3C5F
 
 | | Structure of a binary complex of the R517A Pol lambda mutant | | Descriptor: | DNA (5'-D(*DCP*DAP*DGP*DTP*DAP*DC)-3'), DNA (5'-D(*DCP*DGP*DGP*DCP*DCP*DGP*DTP*DAP*DCP*DTP*DG)-3'), DNA (5'-D(P*DGP*DCP*DCP*DG)-3'), ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Foley, M.C, Pedersen, L.C, Schlick, T, Kunkel, T.A. | | Deposit date: | 2008-01-31 | | Release date: | 2008-09-02 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate-induced DNA strand misalignment during catalytic cycling by DNA polymerase lambda.
Embo Rep., 9, 2008
|
|
8DB4
 
 | |
4OUO
 
 | | anti-Bla g 1 scFv | | Descriptor: | CHLORIDE ION, SULFATE ION, anti Bla g 1 scFv | | Authors: | Mueller, G.A, Ankney, J.A, Glesner, J, Khurana, T, Edwards, L.L, Pedersen, L.C, Perera, L, Slater, J.E, Pomes, A, London, R.E. | | Deposit date: | 2014-02-18 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of an anti-Bla g 1 scFv: Epitope mapping and cross-reactivity.
Mol.Immunol., 59, 2014
|
|
4QH0
 
 | | Crystal structure of NucA from Streptococcus agalactiae with magnesium ion bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-entry nuclease (Competence-specific nuclease), ... | | Authors: | Moon, A.F, Gaudu, P, Pedersen, L.C. | | Deposit date: | 2014-05-26 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of the virulence factor nuclease A from Streptococcus agalactiae.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3OB4
 
 | | MBP-fusion protein of the major peanut allergen Ara h 2 | | Descriptor: | CHLORIDE ION, Maltose ABC transporter periplasmic protein,Arah 2, SULFATE ION, ... | | Authors: | Mueller, G.A, Gosavi, R.A, Moon, A.F, London, R.E, Pedersen, L.C. | | Deposit date: | 2010-08-06 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Ara h 2: crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity.
Allergy, 66, 2011
|
|
3MQ1
 
 | | Crystal Structure of Dust Mite Allergen Der p 5 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Mite allergen Der p 5, ... | | Authors: | Mueller, G.A, Gosavi, R.A, Krahn, J.M, Edwards, L.L, Cuneo, M.J, Glesner, J, Pomes, A, Chapman, M.D, London, R.E, Pedersen, L.C. | | Deposit date: | 2010-04-27 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Der p 5 crystal structure provides insight into the group 5 dust mite allergens.
J.Biol.Chem., 285, 2010
|
|
3MBY
 
 | | Ternary complex of DNA Polymerase BETA with template base A and 8oxodGTP in the active site with a dideoxy terminated primer | | Descriptor: | 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Hou, E.W, Pedersen, L.C, Prasad, R, Wilson, S.H. | | Deposit date: | 2010-03-26 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenic conformation of 8-oxo-7,8-dihydro-2'-dGTP in the confines of a DNA polymerase active site.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3OWV
 
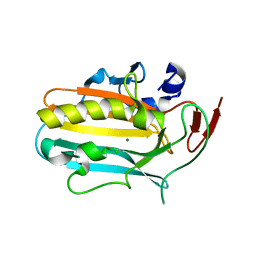 | | Structural insights into catalytic and substrate binding mechanisms of the strategic EndA nuclease from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, DNA-entry nuclease, MAGNESIUM ION | | Authors: | Moon, A.F, Midon, M, Meiss, G, Pingoud, A.M, London, R.E, Pedersen, L.C. | | Deposit date: | 2010-09-20 | | Release date: | 2010-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into catalytic and substrate binding mechanisms of the strategic EndA nuclease from Streptococcus pneumoniae.
Nucleic Acids Res., 39, 2011
|
|
3PML
 
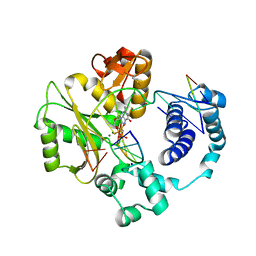 | | crystal structure of a polymerase lambda variant with a dGTP analog opposite a templating T | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]guanosine, 5'-D(*CP*AP*GP*TP*AP*C)-3', 5'-D(*CP*GP*GP*CP*TP*GP*TP*AP*CP*TP*G)-3', ... | | Authors: | Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2010-11-17 | | Release date: | 2011-01-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Replication infidelity via a mismatch with Watson-Crick geometry.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PNC
 
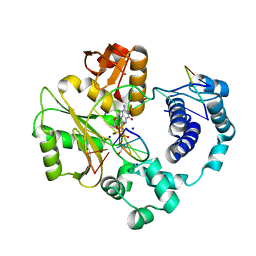 | | Ternary crystal structure of a polymerase lambda variant with a GT mispair at the primer terminus and sodium at catalytic metal site | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]guanosine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*CP*AP*GP*TP*AP*G)-3', ... | | Authors: | Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2010-11-18 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Replication infidelity via a mismatch with Watson-Crick geometry.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PMN
 
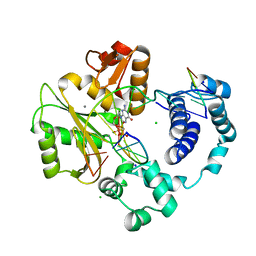 | | ternary crystal structure of polymerase lambda variant with a GT mispair at the primer terminus with Mn2+ in the active site | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]guanosine, 5'-D(*CP*AP*GP*TP*AP*G)-3', 5'-D(*CP*GP*GP*CP*CP*TP*TP*AP*CP*TP*G)-3', ... | | Authors: | Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2010-11-17 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Replication infidelity via a mismatch with Watson-Crick geometry.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1BO6
 
 | |
3UAN
 
 | | Crystal structure of 3-O-sulfotransferase (3-OST-1) with bound PAP and heptasaccharide substrate | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Moon, A.F, Xu, Y, Woody, S.M, Krahn, J.M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2011-10-21 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Dissecting the substrate recognition of 3-O-sulfotransferase for the biosynthesis of anticoagulant heparin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6TYW
 
 | |
6TYZ
 
 | |
6TYV
 
 | |
