6KZE
 
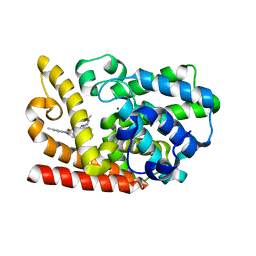 | | The crystal structue of PDE10A complexed with 4d | | Descriptor: | 8-[(E)-2-[5-methyl-1-[3-[3-(4-methylpiperazin-1-yl)propoxy]phenyl]benzimidazol-2-yl]ethenyl]quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Yang, Y, Zhang, S, Zhou, Q, Huang, Y.-Y, Guo, L, Luo, H.-B. | | Deposit date: | 2019-09-24 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.50003481 Å) | | Cite: | Novel Potent and Highly Selective Benzoimidazole-based Phosphodiesterase 10 Inhibitors with Improved Solubility and Pharmacokinetic Properties for the Treatment of Pulmonary Arterial Hypertension
To Be Published
|
|
4WYH
 
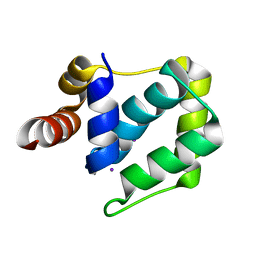 | |
7D77
 
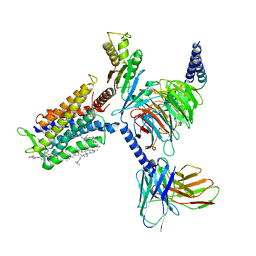 | | Cryo-EM structure of the cortisol-bound adhesion receptor GPR97-Go complex | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | Authors: | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
8W4Q
 
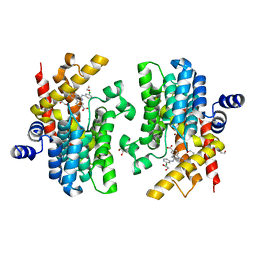 | | Crystal structure of PDE4D complexed with CX-4945 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8W4R
 
 | | Crystal structure of PDE4D complexed with CVT-313 | | Descriptor: | 1,2-ETHANEDIOL, 2,2'-{[6-{[(4-methoxyphenyl)methyl]amino}-9-(propan-2-yl)-9H-purin-2-yl]azanediyl}di(ethan-1-ol), MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8W4S
 
 | | Crystal structure of PDE5A in complex with CVT-313 | | Descriptor: | 2,2'-{[6-{[(4-methoxyphenyl)methyl]amino}-9-(propan-2-yl)-9H-purin-2-yl]azanediyl}di(ethan-1-ol), MAGNESIUM ION, ZINC ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8W4T
 
 | | Crystal structure of PDE5A in complex with a novel inhibitor | | Descriptor: | 2-[bis(2-hydroxyethyl)amino]-6-[(4-methoxyphenyl)methylamino]-9-propan-2-yl-7~{H}-purin-8-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
1NI7
 
 | | NORTHEAST STRUCTURAL GENOMIC CONSORTIUM TARGET ER75 | | Descriptor: | Hypothetical protein ygdK | | Authors: | Liu, G, Chiang, Y, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-12-21 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-quality homology models derived from NMR and X-ray structures of E. coli proteins YgdK and Suf E suggest that all members of the YgdK/Suf E protein family are enhancers of cysteine desulfurases.
Protein Sci., 14, 2005
|
|
1ALQ
 
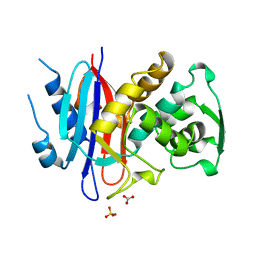 | |
8XKL
 
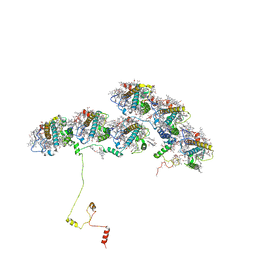 | | Structure of ACPII-CCPII from cryptophyte algae | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, ... | | Authors: | Li, X.Y, Mao, Z.Y, Shen, J.R, Han, G.Y. | | Deposit date: | 2023-12-23 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structure and distinct supramolecular organization of a PSII-ACPII dimer from a cryptophyte alga Chroomonas placoidea.
Nat Commun, 15, 2024
|
|
4Y86
 
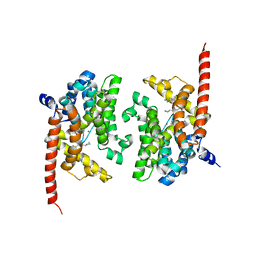 | | Crystal structure of PDE9 in complex with racemic inhibitor C33 | | Descriptor: | 6-{[(1R)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, 6-{[(1S)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, ... | | Authors: | Huang, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-09-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Asymmetry of Phosphodiesterase-9A and a Unique Pocket for Selective Binding of a Potent Enantiomeric Inhibitor.
Mol.Pharmacol., 88, 2015
|
|
7D76
 
 | | Cryo-EM structure of the beclomethasone-bound adhesion receptor GPR97-Go complex | | Descriptor: | (8~{S},9~{R},10~{S},11~{S},13~{S},14~{S},16~{S},17~{R})-9-chloranyl-10,13,16-trimethyl-11,17-bis(oxidanyl)-17-(2-oxidanylethanoyl)-6,7,8,11,12,14,15,16-octahydrocyclopenta[a]phenanthren-3-one, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | Authors: | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
7WQZ
 
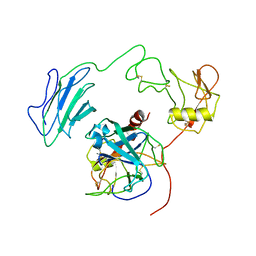 | | Structure of Active-mutEP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain | | Authors: | Yang, X.L, Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-10-26 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
7WQW
 
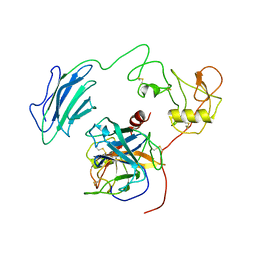 | | Structure of Active-EP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain | | Authors: | Yang, X.L, Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-10-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
7WQX
 
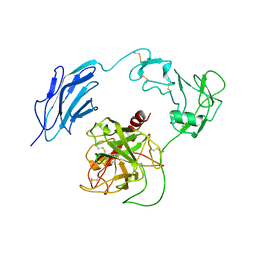 | | Structure of Inactive-EP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Enteropeptidase | | Authors: | Yang, X.L, Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-10-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
7WR7
 
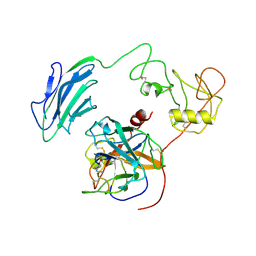 | | Structure of Inhibited-EP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, Enteropeptidase catalytic light chain, ... | | Authors: | Yang, X.L, Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
4Y87
 
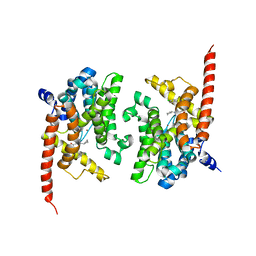 | | Crystal structure of phosphodiesterase 9 in complex with (R)-C33 (6-{[(1R)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one) | | Descriptor: | 6-{[(1R)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Huang, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-09-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Asymmetry of Phosphodiesterase-9A and a Unique Pocket for Selective Binding of a Potent Enantiomeric Inhibitor.
Mol.Pharmacol., 88, 2015
|
|
4Y8C
 
 | | Crystal structure of phosphodiesterase 9 in complex with (S)-C33 | | Descriptor: | 6-{[(1S)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Huang, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-09-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Asymmetry of Phosphodiesterase-9A and a Unique Pocket for Selective Binding of a Potent Enantiomeric Inhibitor.
Mol.Pharmacol., 88, 2015
|
|
7X1N
 
 | | Crystal structure of MEF2D-MRE complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*G)-3'), DNA (5'-D(P*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), Myocyte enhancer factor 2D/deleted in azoospermia associated protein 1 fusion protein | | Authors: | Zhang, H, Zhang, M, Wang, Q.Q, Chen, Z, Chen, S.J, Meng, G. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.315 Å) | | Cite: | Functional, structural, and molecular characterizations of the leukemogenic driver MEF2D-HNRNPUL1 fusion.
Blood, 140, 2022
|
|
7EW1
 
 | | Cryo-EM structure of siponimod -bound Sphingosine-1-phosphate receptor 5 in complex with Gi protein | | Descriptor: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Y, Jia, G.W, Shao, Z.H, Su, Z.M. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
7EW7
 
 | | Cryo-EM structure of SEW2871-bound Sphingosine-1-phosphate receptor 1 in complex with Gi protein | | Descriptor: | 5-[4-phenyl-5-(trifluoromethyl)thiophen-2-yl]-3-[3-(trifluoromethyl)phenyl]-1,2,4-oxadiazole, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jia, G.W, Yuan, Y, Su, Z.M, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
7EVZ
 
 | | Cryo-EM structure of cenerimod -bound Sphingosine-1-phosphate receptor 1 in complex with Gi protein | | Descriptor: | (2~{S})-3-[4-[5-(2-cyclopentyl-6-methoxy-pyridin-4-yl)-1,2,4-oxadiazol-3-yl]-2-ethyl-6-methyl-phenoxy]propane-1,2-diol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Y, Jia, G.W, Shao, Z.H, Su, Z.M. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
7EW0
 
 | | Cryo-EM structure of ozanimod -bound Sphingosine-1-phosphate receptor 1 in complex with Gi protein | | Descriptor: | 5-[3-[(1~{S})-1-(2-hydroxyethylamino)-2,3-dihydro-1~{H}-inden-4-yl]-1,2,4-oxadiazol-5-yl]-2-propan-2-yloxy-benzenecarbonitrile, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Y, Jia, G.W, Su, Z.M, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
7EVY
 
 | | Cryo-EM structure of siponimod -bound Sphingosine-1-phosphate receptor 1 in complex with Gi protein | | Descriptor: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jia, G.W, Yuan, Y, Su, Z.M, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
4HK7
 
 | | Crystal structure of Cordyceps militaris IDCase in complex with uracil | | Descriptor: | URACIL, Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Zhu, J, Ding, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
