8J8G
 
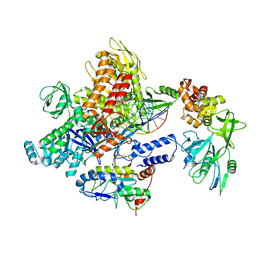 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 in complex with a DNA duplex and cidofovir diphosphate | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*GP*CP*TP*GP*CP*TP*AP*TP*GP*AP*GP*AP*TP*TP*AP*AP*GP*TP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*GP*AP*TP*AP*AP*CP*TP*TP*AP*AP*TP*CP*TP*CP*AP*CP*AP*TP*AP*GP*CP*AP*GP*CP*T)-3'), ... | | Authors: | Xu, Y, Wu, Y, Wu, X, Zhang, Y, Yang, Y, Li, D, Yang, B, Gao, K, Zhang, Z, Dong, C. | | Deposit date: | 2023-05-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural basis of human mpox viral DNA replication inhibition by brincidofovir and cidofovir.
Int.J.Biol.Macromol., 270, 2024
|
|
8J8F
 
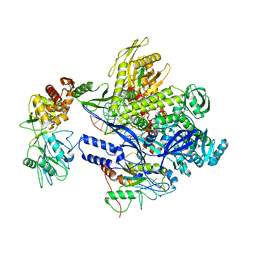 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 in complex with a DNA duplex and dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*AP*GP*CP*TP*GP*CP*TP*AP*TP*GP*AP*GP*AP*TP*TP*AP*AP*GP*TP*TP*AP*T)-3'), ... | | Authors: | Xu, Y, Wu, Y, Wu, X, Zhang, Y, Yang, Y, Li, D, Yang, B, Gao, K, Zhang, Z, Dong, C. | | Deposit date: | 2023-05-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of human mpox viral DNA replication inhibition by brincidofovir and cidofovir.
Int.J.Biol.Macromol., 270, 2024
|
|
8JCB
 
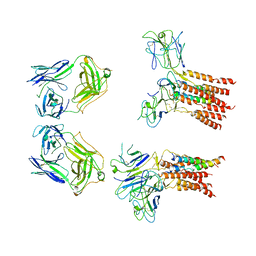 | | Vgamma5 Vdelta1 T cell receptor complex | | Descriptor: | T cell receptor delta variable 1,T cell receptor delta constant, T cell receptor gamma variable 5,T cell receptor gamma constant 1, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
6VUA
 
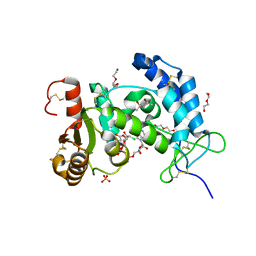 | | X-ray structure of human CD38 catalytic domain with 2'-Cl-araNAD+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dai, Z, Zhang, X.N, Nasertorabi, F, Han, G.W, Stevens, R.C, Zhang, Y. | | Deposit date: | 2020-02-14 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis of site-specific antibody-drug conjugates by ADP-ribosyl cyclases.
Sci Adv, 6, 2020
|
|
2FF0
 
 | | Solution Structure of Steroidogenic Factor 1 DNA Binding Domain Bound to its Target Sequence in the Inhibin alpha-subunit Promoter | | Descriptor: | CTGTGGCCCTGAGCC, GGCTCAGGGCCACAG, Steroidogenic factor 1, ... | | Authors: | Little, T.H, Zhang, Y, Matulis, C.K, Weck, J, Zhang, Z, Ramachandran, A, Mayo, K.E, Radhakrishnan, I. | | Deposit date: | 2005-12-17 | | Release date: | 2006-04-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific deoxyribonucleic Acid (DNA) recognition by steroidogenic factor 1: a helix at the carboxy terminus of the DNA binding domain is necessary for complex stability.
Mol.Endocrinol., 20, 2006
|
|
7XZO
 
 | | Formate-tetrahydrofolate ligase in complex with ATP | | Descriptor: | (R,R)-2,3-BUTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Formate--tetrahydrofolate ligase, ... | | Authors: | Fang, C.L, Zhang, Y. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of FtfL as a novel target of berberine in intestinal bacteria.
Bmc Biol., 21, 2023
|
|
7XZN
 
 | |
7XZP
 
 | |
8K14
 
 | | X-ray crystal structure of 18a in BRD4(1) | | Descriptor: | 4-[8-methoxy-2-methyl-1-(1-phenylethyl)imidazo[4,5-c]quinolin-7-yl]-3,5-dimethyl-1,2-oxazole, Bromodomain-containing protein 4 | | Authors: | Xu, H, Shen, H, Zhang, Y, Xu, Y, Li, R. | | Deposit date: | 2023-07-10 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Discovery of (R)-4-(8-methoxy-2-methyl-1-(1-phenylethy)-1H-imidazo[4,5-c]quinnolin-7-yl)-3,5-dimethylisoxazole as a potent and selective BET inhibitor for treatment of acute myeloid leukemia (AML) guided by FEP calculation.
Eur.J.Med.Chem., 263, 2024
|
|
5Y1T
 
 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,3-dimethylbenzyl)ureido)-N-hydroxy-4-methylpentanamide | | Descriptor: | (2S)-2-[(2,3-dimethylphenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Development of peptidomimetic hydroxamates as PfA-M1 and PfA-M17 dual inhibitors: Biological evaluation and structural characterization by cocrystallization
Chin.Chem.Lett., 33, 2022
|
|
5Y1K
 
 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2-chlorobenzyl)ureido)-N-hydroxy-4-methylpentanamide | | Descriptor: | (2S)-2-[(2-chlorophenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Development of peptidomimetic hydroxamates as PfA-M1 and PfA-M17 dual inhibitors: Biological evaluation and structural characterization by cocrystallization
Chin.Chem.Lett., 33, 2022
|
|
2MK6
 
 | |
5Y1Q
 
 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(3-chlorobenzyl)ureido)-N-hydroxy-4-methylpentanamide | | Descriptor: | (2S)-2-[(3-chlorophenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, M1 family aminopeptidase, MAGNESIUM ION, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Development of peptidomimetic hydroxamates as PfA-M1 and PfA-M17 dual inhibitors: Biological evaluation and structural characterization by cocrystallization
Chin.Chem.Lett., 33, 2022
|
|
7YL2
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004 | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, N-(1-ethyl-2-oxidanylidene-3H-indol-5-yl)cyclohexanesulfonamide, ... | | Authors: | Huang, Y, Wei, A, Dong, R, Xu, H, Zhang, C, Chen, Z, Li, J, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-07-25 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004
To Be Published
|
|
5XM7
 
 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-((2S,3R)-3-amino-2-hydroxy-4-phenylbutanamido)-N-hydroxy-4-methylpentanamide | | Descriptor: | (2S)-4-methyl-N-[(1R)-2-(oxidanylamino)-2-oxidanylidene-1-phenyl-ethyl]-2-[(phenylmethyl)carbamoylamino]pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-05-12 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Development of peptidomimetic hydroxamates as PfA-M1 and PfA-M17 dual inhibitors: Biological evaluation and structural characterization by cocrystallization
Chin.Chem.Lett., 33, 2022
|
|
5X57
 
 | | Structure of GAR domain of ACF7 | | Descriptor: | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5, NICKEL (II) ION | | Authors: | Yang, F, Wang, T, Zhang, Y, Wu, X.Y. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | ACF7 regulates inflammatory colitis and intestinal wound response by orchestrating tight junction dynamics.
Nat Commun, 8, 2017
|
|
8K6L
 
 | | Cryo-EM structure of human OATP1B1 in complex with DCF | | Descriptor: | 2',7'-bis(chloranyl)-3',6'-bis(oxidanyl)spiro[2-benzofuran-3,9'-xanthene]-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shan, Z, Yang, X, Zhang, Y. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
4OHR
 
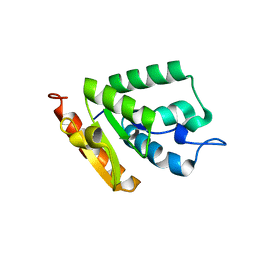 | | Crystal structure of MilB from Streptomyces rimofaciens | | Descriptor: | CMP/hydroxymethyl CMP hydrolase | | Authors: | Zhao, G, Zhang, Y, Liu, G, Wu, G, He, X. | | Deposit date: | 2014-01-17 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the N-glycosidase MilB in complex with hydroxymethyl CMP reveals its Arg23 specifically recognizes the substrate and controls its entry
Nucleic Acids Res., 42, 2014
|
|
4OHB
 
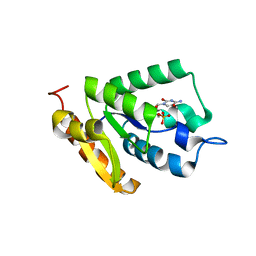 | | Crystal structure of MilB E103A in complex with 5-hydroxymethylcytidine 5'-monophosphate (hmCMP) from Streptomyces rimofaciens | | Descriptor: | 5-(hydroxymethyl)cytidine 5'-(dihydrogen phosphate), CMP/hydroxymethyl CMP hydrolase | | Authors: | Zhao, G, Zhang, Y, Liu, G, Wu, G, He, X. | | Deposit date: | 2014-01-17 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the N-glycosidase MilB in complex with hydroxymethyl CMP reveals its Arg23 specifically recognizes the substrate and controls its entry
Nucleic Acids Res., 42, 2014
|
|
3UAW
 
 | | Crystal structure of adenosine phosphorylase from Bacillus cereus complexed with adenosine | | Descriptor: | ADENOSINE, GLYCEROL, Purine nucleoside phosphorylase deoD-type, ... | | Authors: | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TKJ
 
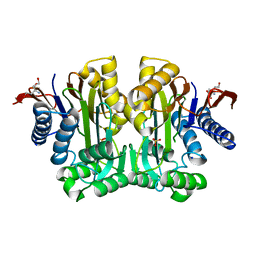 | | Crystal Structure of Human Asparaginase-like Protein 1 Thr168Ala | | Descriptor: | L-asparaginase, SODIUM ION, SULFATE ION, ... | | Authors: | Li, W.Z, Yogesha, S.D, Liu, J, Zhang, Y. | | Deposit date: | 2011-08-26 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Uncoupling Intramolecular Processing and Substrate Hydrolysis in the N-Terminal Nucleophile Hydrolase hASRGL1 by Circular Permutation.
Acs Chem.Biol., 7, 2012
|
|
3UAV
 
 | | Crystal structure of adenosine phosphorylase from Bacillus cereus | | Descriptor: | GLYCEROL, Purine nucleoside phosphorylase deoD-type, SULFATE ION | | Authors: | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3UAX
 
 | | Crystal structure of adenosine phosphorylase from Bacillus cereus complexed with inosine | | Descriptor: | GLYCEROL, INOSINE, Purine nucleoside phosphorylase deoD-type, ... | | Authors: | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3V8T
 
 | |
3VF8
 
 | |
