4IJD
 
 | | Crystal structure of methyltransferase domain of human PR domain-containing protein 9 | | Descriptor: | Histone-lysine N-methyltransferase PRDM9, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Dong, A, Dombrovski, L, Li, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-21 | | Release date: | 2013-02-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of methyltransferase domain of human PR domain-containing protein 9
To be Published
|
|
4JXM
 
 | | Crystal structure of RRP9 WD40 repeats | | Descriptor: | U3 small nucleolar RNA-interacting protein 2, UNKNOWN ATOM OR ION | | Authors: | Wu, X, Tempel, W, Xu, C, El Bakkouri, M, He, H, Seitova, A, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-28 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of RRP9 WD40 repeats
To be Published
|
|
7U9I
 
 | | Co-crystal structure of human CARM1 in complex with MT556 inhibitor | | Descriptor: | 7-[5-S-(4-{[(4-ethylpyridin-3-yl)methyl]amino}butyl)-5-thio-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Histone-arginine methyltransferase CARM1, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Perveen, S, Dong, A, Hutchinson, A, Seitova, A, Gibson, E, Hajian, T, Li, Y, Gao, Y.D, Schneider, S, Siliphaivanh, P, Sloman, D, Nicholson, B, Fischer, C, Hicks, J, Vedadi, M, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal structure of human CARM1 in complex with MT556 inhibitor
To Be Published
|
|
7UNK
 
 | | Structure of Importin-4 bound to the H3-H4-ASF1 histone-histone chaperone complex | | Descriptor: | Histone H3, Histone H4, Histone chaperone, ... | | Authors: | Bernardes, N.E, Chook, Y.M, Fung, H.Y.J, Chen, Z, Li, Y. | | Deposit date: | 2022-04-11 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of IMPORTIN-4 bound to the H3-H4-ASF1 histone-histone chaperone complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5FAI
 
 | | EMG1 N1-Specific Pseudouridine Methyltransferase | | Descriptor: | CITRIC ACID, Ribosomal RNA small subunit methyltransferase NEP1, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | DONG, A, ZENG, H, LI, Y, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | EMG1 N1-Specific Pseudouridine Methyltransferase
to be published
|
|
8WA3
 
 | | Cryo-EM structure of peptide free and Gs-coupled GIPR | | Descriptor: | Gastric inhibitory polypeptide receptor,Fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,O-antigen polymerase, ... | | Authors: | Cong, Z.T, Zhao, F.H, Li, Y, Luo, G, Zhou, Q.T, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-09-06 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Molecular features of the ligand-free GLP-1R, GCGR and GIPR in complex with G s proteins.
Cell Discov, 10, 2024
|
|
8WG8
 
 | | Cryo-EM structures of peptide free and Gs-coupled GCGR | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cong, Z.T, Zhao, F.H, Li, Y, Luo, G, Zhou, Q.T, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-09-20 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Molecular features of the ligand-free GLP-1R, GCGR and GIPR in complex with G s proteins.
Cell Discov, 10, 2024
|
|
8WG7
 
 | | Cryo-EM structures of peptide free and Gs-coupled GLP-1R | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cong, Z.T, Zhao, F.H, Li, Y, Luo, G, Zhou, Q.T, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-09-20 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Molecular features of the ligand-free GLP-1R, GCGR and GIPR in complex with G s proteins.
Cell Discov, 10, 2024
|
|
4K12
 
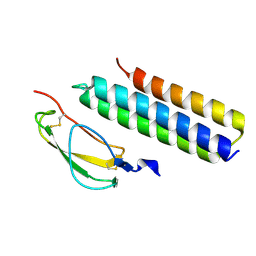 | | Structural Basis for Host Specificity of Factor H Binding by Streptococcus pneumoniae | | Descriptor: | Choline binding protein A, Complement factor H | | Authors: | Liu, A, Achila, D, Banerjee, R, Martinez-Hackert, E, Li, Y, Yan, H. | | Deposit date: | 2013-04-04 | | Release date: | 2014-04-09 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.079 Å) | | Cite: | Structural determinants of host specificity of complement Factor H recruitment by Streptococcus pneumoniae.
Biochem.J., 465, 2015
|
|
4KY1
 
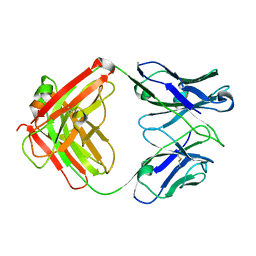 | | humanized HP1/2 Fab | | Descriptor: | IMMUNOGLOBULIN IGG1 FAB, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Arndt, J.W, Hanf, K.J.M, Lugovskoy, A, Chen, L.L, Jarpe, M, Boriack-Sjodin, A, Li, Y, van Vlijmen, H, Pepinsky, B, Taylor, F, Silvian, L, Taveras, A. | | Deposit date: | 2013-05-28 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Antibody humanization by redesign of complementarity-determining region residues proximate to the acceptor framework.
METHODS (SAN DIEGO), 65, 2014
|
|
5FRS
 
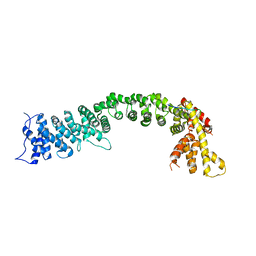 | | Structure of the Pds5-Scc1 complex and implications for cohesin function | | Descriptor: | SISTER CHROMATID COHESION PROTEIN 1, SISTER CHROMATID COHESION PROTEIN PDS5 | | Authors: | Muir, K.W, Kschonsak, M, Li, Y, Metz, J, Haering, C.H, Panne, D. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.073 Å) | | Cite: | Structure of the Pds5-Scc1 Complex and Implications for Cohesin Function
Cell Rep., 14, 2016
|
|
5FRP
 
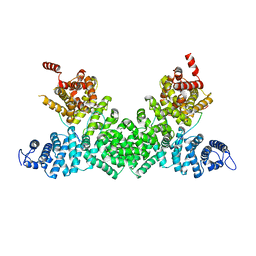 | | Structure of the Pds5-Scc1 complex and implications for cohesin function | | Descriptor: | MCD1-LIKE PROTEIN, SISTER CHROMATID COHESION PROTEIN PDS5 | | Authors: | Muir, K.W, Kschonsak, M, Li, Y, Metz, J, Haering, C.H, Panne, D. | | Deposit date: | 2015-12-21 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | Structure of the Pds5-Scc1 Complex and Implications for Cohesin Function
Cell Rep., 14, 2016
|
|
5GJ4
 
 | |
5H43
 
 | |
4J4Q
 
 | | Crystal structure of active conformation of GPCR opsin stabilized by octylglucoside | | Descriptor: | ACETATE ION, Guanine nucleotide-binding protein G(t) subunit alpha-1, PALMITIC ACID, ... | | Authors: | Park, J.H, Morizumi, T, Li, Y, Hong, J.E, Pai, E.F, Hofmann, K.P, Choe, H.W, Ernst, O.P. | | Deposit date: | 2013-02-07 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Opsin, a structural model for olfactory receptors?
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
5W1Y
 
 | | SETD8 in complex with a covalent inhibitor | | Descriptor: | 2-(4-methylpiperazin-1-yl)-3-(phenylsulfanyl)naphthalene-1,4-dione, N-lysine methyltransferase KMT5A, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Yu, W, Li, Y, Blum, G, Luo, M, Pittella-Silva, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-05 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SETD8 in complex with a covalent inhibitor
to be published
|
|
5VLQ
 
 | | Structure of the TTLL3 Glycylase | | Descriptor: | LOC100158544 protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION | | Authors: | Garnham, C.P, Yu, I, Li, Y, Roll-Mecak, A. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Crystal structure of tubulin tyrosine ligase-like 3 reveals essential architectural elements unique to tubulin monoglycylases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8VIS
 
 | | Human TMPRSS11D complexed with a disulfide-linked autoinhibitory DDDDK peptide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fraser, B.J, Dong, A, Ilyassov, O, Kenney, T, Li, Y.Y, Seitova, A, Li, Y, Hejazi, Z, Edwards, A, Benard, F, Arrowsmith, C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Human TMPRSS11D complexed with a disulfide-linked autoinhibitory DDDDK peptide
To Be Published
|
|
6LZ2
 
 | | Crystal structure of a thermostable green fluorescent protein (TGP) with a synthetic nanobody (Sb44) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Cai, H, Yao, H, Li, T, Hutter, C, Tang, Y, Li, Y, Seeger, M, Li, D. | | Deposit date: | 2020-02-17 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | An improved fluorescent tag and its nanobodies for membrane protein expression, stability assay, and purification.
Commun Biol, 3, 2020
|
|
5Y5N
 
 | | Crystal structure of human Sirtuin 2 in complex with a selective inhibitor | | Descriptor: | 2-[[3-(2-phenylethoxy)phenyl]amino]benzamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | Authors: | Mellini, P, Itoh, Y, Tsumoto, H, Li, Y, Suzuki, M, Tokuda, N, Kakizawa, T, Miura, Y, Takeuchi, J, Lahtela-Kakkonen, M, Suzuki, T. | | Deposit date: | 2017-08-09 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent mechanism-based sirtuin-2-selective inhibition by anin situ-generated occupant of the substrate-binding site, "selectivity pocket" and NAD+-binding site.
Chem Sci, 8, 2017
|
|
5X29
 
 | |
5XI4
 
 | | BRD4 bound with compound Bdi4 | | Descriptor: | (3~{S})-4-cyclopropyl-1,3-dimethyl-6-[[(1~{S})-1-(4-methylphenyl)ethyl]amino]-3~{H}-quinoxalin-2-one, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D, Li, Y. | | Deposit date: | 2017-04-25 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.486 Å) | | Cite: | BRD4 bound with compound Bdi4
To Be Published
|
|
5XI3
 
 | | BRD4 bound with compound Bdi3 | | Descriptor: | (3~{R})-4-cyclopropyl-1,3-dimethyl-6-[[(1~{R})-1-phenylethyl]amino]-3~{H}-quinoxalin-2-one, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D, Li, Y. | | Deposit date: | 2017-04-25 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.674 Å) | | Cite: | BRD4 bound with compound Bdi3
To Be Published
|
|
8T9A
 
 | | CryoEM structure of human DDB1-DCAF12 in complex with MAGEA3 | | Descriptor: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, Melanoma-associated antigen 3 | | Authors: | Duda, D, Righetto, G, Li, Y, Loppnau, P, Seitova, A, Santhakumar, V, Halabelian, L, Yin, Y. | | Deposit date: | 2023-06-23 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | CryoEM structure of human DDB1-DCAF12 in complex with MAGEA3
To Be Published
|
|
8Y6P
 
 | |
