5Y5N
 
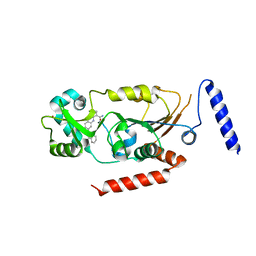 | | Crystal structure of human Sirtuin 2 in complex with a selective inhibitor | | Descriptor: | 2-[[3-(2-phenylethoxy)phenyl]amino]benzamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | Authors: | Mellini, P, Itoh, Y, Tsumoto, H, Li, Y, Suzuki, M, Tokuda, N, Kakizawa, T, Miura, Y, Takeuchi, J, Lahtela-Kakkonen, M, Suzuki, T. | | Deposit date: | 2017-08-09 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent mechanism-based sirtuin-2-selective inhibition by anin situ-generated occupant of the substrate-binding site, "selectivity pocket" and NAD+-binding site.
Chem Sci, 8, 2017
|
|
5XN9
 
 | | Crystal structure of a secretary abundant heat soluble (SAHS) protein from Ramazzottius varieornatus (from monomer sample) | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Fukuda, Y, Miura, Y, Mizohata, E, Inoue, T. | | Deposit date: | 2017-05-19 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structural insights into a secretory abundant heat-soluble protein from an anhydrobiotic tardigrade, Ramazzottius varieornatus
FEBS Lett., 591, 2017
|
|
5XNA
 
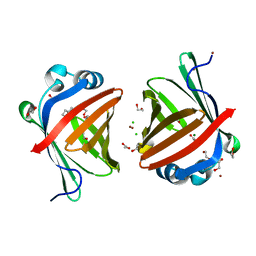 | | Crystal structure of a secretary abundant heat soluble (SAHS) protein from Ramazzottius varieornatus (from dimer sample) | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Fukuda, Y, Miura, Y, Mizohata, E, Inoue, T. | | Deposit date: | 2017-05-19 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural insights into a secretory abundant heat-soluble protein from an anhydrobiotic tardigrade, Ramazzottius varieornatus
FEBS Lett., 591, 2017
|
|
3VGB
 
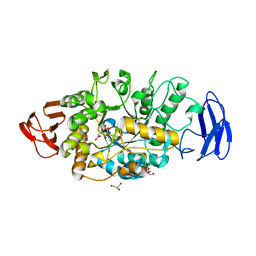 | | Crystal structure of glycosyltrehalose trehalohydrolase (GTHase) from Sulfolobus solfataricus KM1 | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGE
 
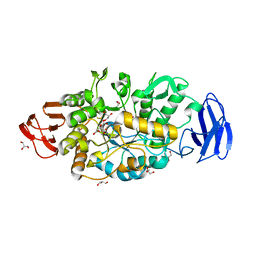 | | Crystal structure of glycosyltrehalose trehalohydrolase (D252S) | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGD
 
 | | Ctystal structure of glycosyltrehalose trehalohydrolase (D252E) | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGF
 
 | | Crystal structure of glycosyltrehalose trehalohydrolase (D252S) complexed with maltotriosyltrehalose | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase, ... | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGH
 
 | | Crystal structure of glycosyltrehalose trehalohydrolase (E283Q) complexed with maltotriosyltrehalose | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase, ... | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGG
 
 | | Crystal structure of glycosyltrehalose trehalohydrolase (E283Q) complexed with maltoheptaose | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase, ... | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
