7DSJ
 
 | |
7DSM
 
 | |
7DSP
 
 | |
7DSR
 
 | |
7DSO
 
 | |
9IVC
 
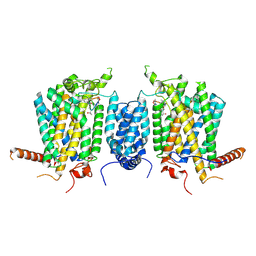 | | Cryo-EM structure of AbA-bound Aur1-Kei1 complex | | Descriptor: | Aureobasidin A, Inositol phosphorylceramide synthase catalytic subunit AUR1, Inositol phosphorylceramide synthase regulatory subunit KEI1 | | Authors: | Xie, T, Wu, X, Gong, X. | | Deposit date: | 2024-07-23 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Mechanisms of aureobasidin A inhibition and drug resistance in a fungal IPC synthase complex.
Nat Commun, 16, 2025
|
|
9LHJ
 
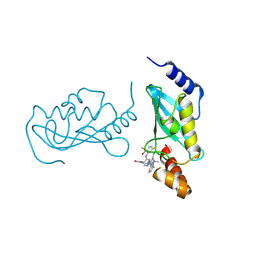 | | UBE2N/UBE2V2 complexed with a covalent inhibitor | | Descriptor: | Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 2 | | Authors: | Li, S, Wu, X, Zhou, L, Lu, X. | | Deposit date: | 2025-01-12 | | Release date: | 2025-05-14 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Proteome-Wide Data Guides the Discovery of Lysine-Targeting Covalent Inhibitors Using DNA-Encoded Chemical Libraries.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9LG2
 
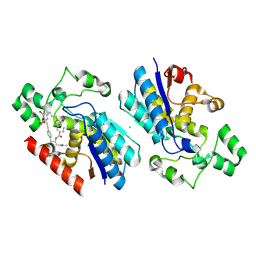 | | Phosphoglycerate mutase 1 complexed with a covalent inhibitor | | Descriptor: | CHLORIDE ION, Phosphoglycerate mutase 1 | | Authors: | Li, S, Wu, X, Zhou, L, Lu, X. | | Deposit date: | 2025-01-09 | | Release date: | 2025-05-14 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Proteome-Wide Data Guides the Discovery of Lysine-Targeting Covalent Inhibitors Using DNA-Encoded Chemical Libraries.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
3TPX
 
 | |
1ZY6
 
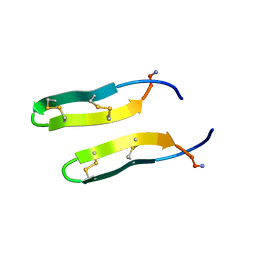 | | Membrane-bound dimer structure of Protegrin-1 (PG-1), a beta-Hairpin Antimicrobial Peptide in Lipid Bilayers from Rotational-Echo Double-Resonance Solid-State NMR | | Descriptor: | Protegrin 1 | | Authors: | Wu, X, Mani, R, Tang, M, Buffy, J.J, Waring, A.J, Sherman, M.A, Hong, M. | | Deposit date: | 2005-06-09 | | Release date: | 2006-06-13 | | Last modified: | 2024-11-13 | | Method: | SOLID-STATE NMR | | Cite: | Membrane-Bound Dimer Structure of a beta-Hairpin Antimicrobial Peptide from Rotational-Echo Double-Resonance Solid-State NMR.
Biochemistry, 45, 2006
|
|
2C2N
 
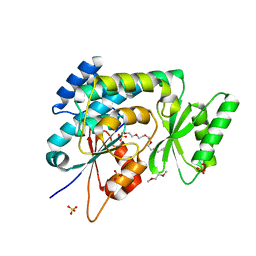 | | Structure of human mitochondrial malonyltransferase | | Descriptor: | 1,2-DIMETHOXYETHANE, 2-(2-ETHOXYETHOXY)ETHANOL, 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, ... | | Authors: | Wu, X, Bunkoczi, G, Smee, C, Arrowsmith, C, Sundstrom, M, Weigelt, J, Edwards, A, von Delft, F, Oppermann, U. | | Deposit date: | 2005-09-29 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Different Specificities of Acyltransferases Associated with the Human Cytosolic and Mitochondrial Fatty Acid Synthases.
Chem.Biol., 16, 2009
|
|
2BEL
 
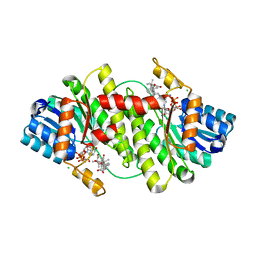 | | Structure of human 11-beta-hydroxysteroid dehydrogenase in complex with NADP and carbenoxolone | | Descriptor: | CARBENOXOLONE, CHLORIDE ION, CORTICOSTEROID 11-BETA-DEHYDROGENASE ISOZYME 1, ... | | Authors: | Kavanagh, K, Wu, X, Svensson, S, Elleby, B, von Delft, F, Debreczeni, J.E, Sharma, S, Bray, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Abrahmsen, L, Oppermann, U. | | Deposit date: | 2004-11-25 | | Release date: | 2004-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The High Resolution Structures of Human, Murine and Guinea Pig 11-Beta-Hydroxysteroid Dehydrogenase Type 1 Reveal Critical Differences in Active Site Architecture
To be Published
|
|
4DU0
 
 | | Crystal structure of human alpha-defensin 1, HNP1 (G17A mutant) | | Descriptor: | CHLORIDE ION, GLYCEROL, Neutrophil defensin 1 | | Authors: | Wu, X, Lu, W, Pazgier, M. | | Deposit date: | 2012-02-21 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Invariant gly residue is important for alpha-defensin folding, dimerization, and function: a case study of the human neutrophil alpha-defensin HNP1
J.Biol.Chem., 287, 2012
|
|
3TNN
 
 | |
3TNM
 
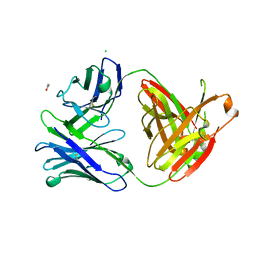 | | Crystal structure of A32 Fab, an ADCC mediating anti-HIV-1 antibody | | Descriptor: | ACETATE ION, CHLORIDE ION, Fab heavy chain of human anti-HIV-1 Env antibody A32, ... | | Authors: | Wu, X, Pazgier, M. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Diverse specificity and effector function among human antibodies to HIV-1 envelope glycoprotein epitopes exposed by CD4 binding.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5DSE
 
 | | Crystal Structure of the TTC7B/Hyccin Complex | | Descriptor: | Hyccin, Tetratricopeptide repeat protein 7B | | Authors: | Wu, X, Baskin, J.M, Reinisch, K.M, De Camilli, P. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The leukodystrophy protein FAM126A (hyccin) regulates PtdIns(4)P synthesis at the plasma membrane.
Nat.Cell Biol., 18, 2016
|
|
4E86
 
 | |
4V0G
 
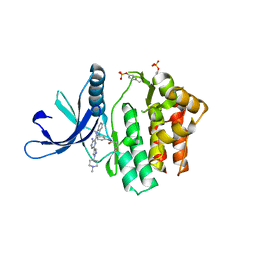 | | JAK3 in complex with a covalent EGFR inhibitor | | Descriptor: | N-[3-(2-{3-amino-6-[1-(1-methylpiperidin-4-yl)-1H-pyrazol-4-yl]pyrazin-2-yl}-1H-benzimidazol-1-yl)phenyl]propanamide, TYROSINE-PROTEIN KINASE JAK3 | | Authors: | Debreczeni, J.E, Hennessy, E.J, Chuaquini, C, Ashton, S, Coclough, N, Cross, D.A.E, Eberlein, C, Gingipalli, L, Klinowska, T.C.M, Orme, J.P, Sha, L, Wu, X. | | Deposit date: | 2014-09-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Utilisation of Structure Based Design to Identify Novel, Irreversible Inhibitors of the Epidermal Growth Factor Receptor (Egfr) Harboring the Gatekeeper T790M Mutation
To be Published
|
|
4W60
 
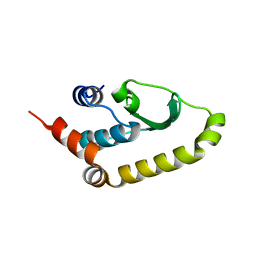 | | The structure of Vaccina virus H7 protein displays A Novel Phosphoinositide binding fold required for membrane biogenesis | | Descriptor: | Late protein H7 | | Authors: | Kolli, S, Meng, X, Wu, X, Shengjuler, D, Cameron, C.E, Xiang, Y, Deng, J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-12-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-function analysis of vaccinia virus h7 protein reveals a novel phosphoinositide binding fold essential for poxvirus replication.
J.Virol., 89, 2015
|
|
5XEZ
 
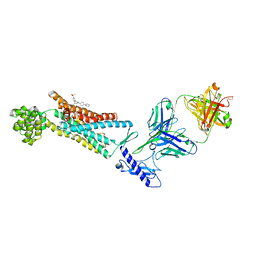 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
4LBB
 
 | |
4LB7
 
 | |
2IC1
 
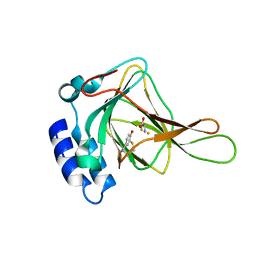 | | Crystal Structure of Human Cysteine Dioxygenase in Complex with Substrate Cysteine | | Descriptor: | CYSTEINE, Cysteine dioxygenase type 1, FE (II) ION | | Authors: | Ye, S, Wu, X, Wei, L, Tang, D, Sun, P, Rao, Z. | | Deposit date: | 2006-09-12 | | Release date: | 2006-12-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An Insight into the Mechanism of Human Cysteine Dioxygenase: KEY ROLES OF THE THIOETHER-BONDED TYROSINE-CYSTEINE COFACTOR.
J.Biol.Chem., 282, 2007
|
|
7EVJ
 
 | | Crystal structure of CBP bromodomain liganded with 9c | | Descriptor: | 3-acetyl-1-((3-(1-cyclopropyl-1H-pyrazol-4-yl)-2-fluoro-5-(hydroxymethyl)phenyl)carbamoyl)indolizin-7-yl dimethylcarbamate, CREB-binding protein, GLYCEROL, ... | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, Y, Zhang, C, Luo, G, Wu, X, Shen, H, Xu, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
4LLW
 
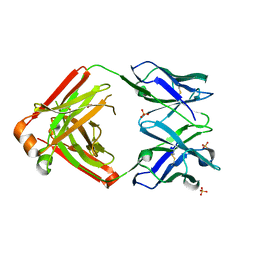 | | Crystal structure of Pertuzumab Clambda Fab with variable domain redesign (VRD2) at 1.95A | | Descriptor: | SULFATE ION, light chain Clambda, mutated Pertuzumab Fab heavy chain | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
