8TVQ
 
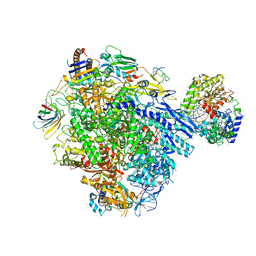 | | Cryo-EM structure of CPD stalled 10-subunit Pol II in complex with Rad26 | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVX
 
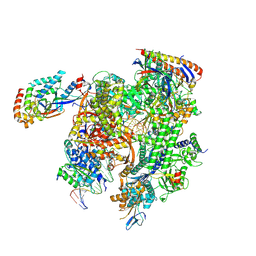 | | Cryo-EM structure of CPD-stalled Pol II (Conformation 2) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVY
 
 | | Cryo-EM structure of CPD lesion containing RNA Polymerase II elongation complex with Rad26 and Elf1 (closed state) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVW
 
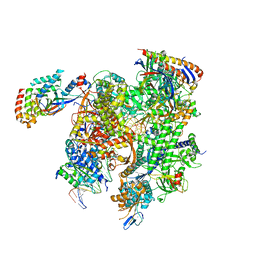 | | Cryo-EM structure of CPD-stalled Pol II (conformation 1) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVS
 
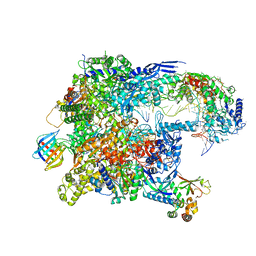 | | Cryo-EM structure of backtracked Pol II in complex with Rad26 | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TUG
 
 | | Cryo-EM structure of CPD-stalled Pol II in complex with Rad26 (engaged state) | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-16 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6LSB
 
 | |
6LS6
 
 | |
3IFU
 
 | | The Crystal Structure of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) Leader Protease Nsp1 | | Descriptor: | Non-structural protein, ZINC ION | | Authors: | Sun, Y, Xue, F, Guo, Y, Ma, M, Lou, Z, Rao, Z. | | Deposit date: | 2009-07-26 | | Release date: | 2009-10-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of porcine reproductive and respiratory syndrome virus leader protease Nsp1alpha
J.Virol., 83, 2009
|
|
6LSD
 
 | |
6M2L
 
 | | Crystal structure of Plasmodium falciparum hexose transporter PfHT1 bound with C3361 | | Descriptor: | (2S,3R,4S,5R,6R)-6-(hydroxymethyl)-4-undec-10-enoxy-oxane-2,3,5-triol, Hexose transporter 1 | | Authors: | Jiang, X, Yuan, Y.Y, Zhang, S, Wang, N, Yan, C.Y, Yan, N. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis for Blocking Sugar Uptake into the Malaria Parasite Plasmodium falciparum.
Cell, 183, 2020
|
|
7X8X
 
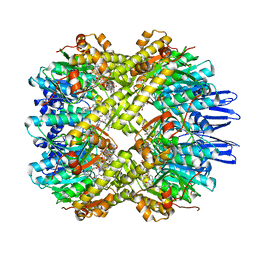 | | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease | | Descriptor: | 4-[1-[3-[4-[(4-fluoranyl-2-methyl-1H-indol-5-yl)oxy]-6-methoxy-quinazolin-7-yl]oxypropyl]piperidin-4-yl]benzamide, 4-[[3,5-bis(fluoranyl)phenyl]methyl]-N-[(4-bromophenyl)methyl]piperazine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit 1, ... | | Authors: | Bao, R, Luo, Y.F, Zhu, Y.B, Yang, Y, Zhou, Y.Z. | | Deposit date: | 2022-03-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Discovery and Mechanistic Study of Novel Mycobacterium tuberculosis ClpP1P2 Inhibitors
J.Med.Chem., 66, 2023
|
|
6M20
 
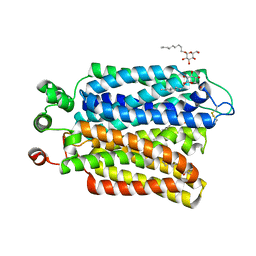 | | Crystal structure of Plasmodium falciparum hexose transporter PfHT1 bound with glucose | | Descriptor: | Hexose transporter 1, beta-D-glucopyranose, nonyl beta-D-glucopyranoside | | Authors: | Jiang, X, Yuan, Y.Y, Zhang, S, Wang, N, Yan, C.Y, Yan, N. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Blocking Sugar Uptake into the Malaria Parasite Plasmodium falciparum.
Cell, 183, 2020
|
|
8I60
 
 | |
8WCO
 
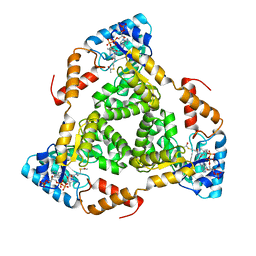 | | (S)-citramalyl-CoA lyase | | Descriptor: | ACETYL COENZYME *A, Probable acyl-CoA lyase beta chain | | Authors: | Huang, Q, Bao, R. | | Deposit date: | 2023-09-13 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterization of itaconyl-CoA hydratase and citramalyl-CoA lyase involved in itaconate metabolism of Pseudomonas aeruginosa.
Structure, 32, 2024
|
|
7BOV
 
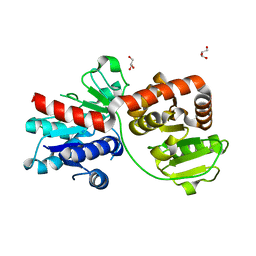 | | The Structure of Bacillus subtilis glycosyltransferase,Bs-YjiC | | Descriptor: | GLYCEROL, SODIUM ION, Uncharacterized UDP-glucosyltransferase YjiC | | Authors: | Zhao, C, Liu, B, Zhao, N.L, Luo, Y.Z, Bao, R. | | Deposit date: | 2020-03-20 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structural and biochemical studies of the glycosyltransferase Bs-YjiC from Bacillus subtilis.
Int.J.Biol.Macromol., 166, 2021
|
|
8KHG
 
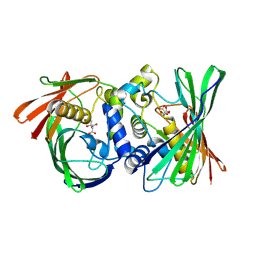 | | Itaconyl-CoA hydratase PaIch | | Descriptor: | CITRIC ACID, Itaconyl-CoA hydratase | | Authors: | Huang, Q, Bao, R. | | Deposit date: | 2023-08-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterization of itaconyl-CoA hydratase and citramalyl-CoA lyase involved in itaconate metabolism of Pseudomonas aeruginosa.
Structure, 32, 2024
|
|
8KHL
 
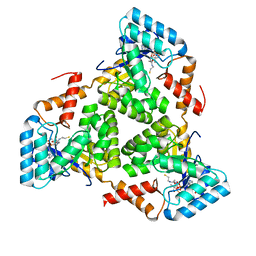 | | (S)-citramalyl-CoA lyase | | Descriptor: | COENZYME A, Probable acyl-CoA lyase beta chain | | Authors: | Huang, Q, Bao, R. | | Deposit date: | 2023-08-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of itaconyl-CoA hydratase and citramalyl-CoA lyase involved in itaconate metabolism of Pseudomonas aeruginosa.
Structure, 32, 2024
|
|
6JAU
 
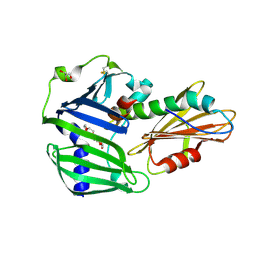 | | The complex structure of Pseudomonas aeruginosa MucA/MucB. | | Descriptor: | CALCIUM ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Li, T, He, L.H, Li, C.C, Liu, L, Peng, C.T, Shen, Y.L, Qin, X.F, Xiao, Q.J, Zhu, Y.B, Song, Y.J, Zhao, N.l, Zhao, C, Yang, J, Mu, X.Y, Huang, Q, Bao, R. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Molecular basis of the lipid-induced MucA-MucB dissociation in Pseudomonas aeruginosa.
Commun Biol, 3, 2020
|
|
5YEI
 
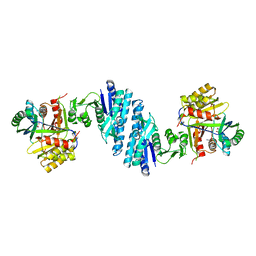 | | Mechanistic insight into the regulation of Pseudomonas aeruginosa aspartate kinase | | Descriptor: | Aspartokinase, GLYCEROL, LYSINE, ... | | Authors: | Li, C, Yang, M, Liu, L, Peng, C, Li, T, He, L, Song, Y, Zhu, Y, Bao, R. | | Deposit date: | 2017-09-17 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into the allosteric regulation of Pseudomonas aeruginosa aspartate kinase.
Biochem.J., 475, 2018
|
|
7CRZ
 
 | | Crystal structure of human glucose transporter GLUT3 bound with C3361 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3R,4S,5R,6R)-6-(hydroxymethyl)-4-undec-10-enoxy-oxane-2,3,5-triol, Solute carrier family 2, ... | | Authors: | Yuan, Y.Y, Zhang, S, Wang, N, Jiang, X, Yan, N. | | Deposit date: | 2020-08-14 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Orthosteric-allosteric dual inhibitors of PfHT1 as selective antimalarial agents.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DEO
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Spike protein S1, ... | | Authors: | Fu, D, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
7DET
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Wang, Y, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
7DEU
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Zhang, Z, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
7CO3
 
 | | HtrA-type protease AlgWS227A with tripeptide | | Descriptor: | AlgW protein, TRP-VAL-PHE | | Authors: | Li, T, Song, Y.J, Bao, R. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis of the Versatile Regulatory Mechanism of HtrA-Type Protease AlgW from Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
