7VBI
 
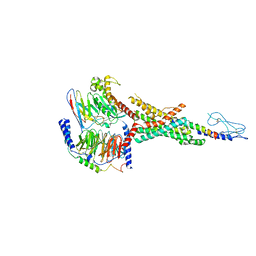 | | Cryo-EM structure of the non-acylated tirzepatide (LY3298176)-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
8FLI
 
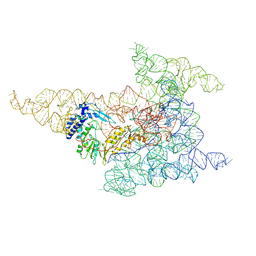 | | Cryo-EM structure of a group II intron immediately before branching | | Descriptor: | Group II Intron, MAGNESIUM ION, Maturase reverse transcriptase | | Authors: | Haack, D.B, Rudolfs, B.G, Zhang, C, Lyumkis, D, Toor, N. | | Deposit date: | 2022-12-21 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of branching during RNA splicing.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HGH
 
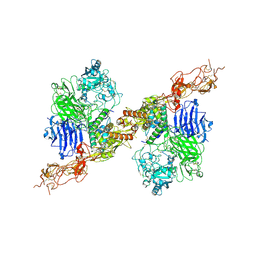 | | Structure of 2:2 PAPP-A.STC2 complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
8HGG
 
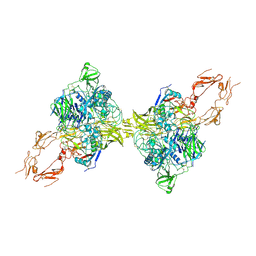 | | Structure of 2:2 PAPP-A.ProMBP complex | | Descriptor: | Bone marrow proteoglycan, Pappalysin-1, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
8HD7
 
 | | The intermediate pre-Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The intermediate pre-Tet-S1 state molecule of co-transcriptional folded G264A mutant Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
8HD6
 
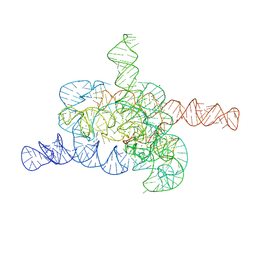 | | The relaxed pre-Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The relaxed pre-Tet-S1 state molecule of co-transcriptional folded G264A mutant Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
8I7N
 
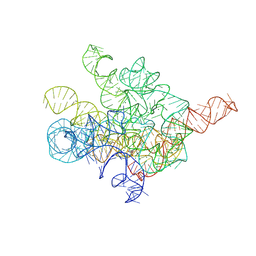 | | The Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Descriptor: | (2R,3R,4S,5R)-2-(2-azanylpurin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol, MAGNESIUM ION, SPERMIDINE, ... | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2023-02-01 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
8JRU
 
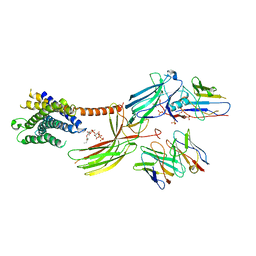 | | Cryo-EM structure of the glucagon receptor bound to beta-arrestin 1 in ligand-free state | | Descriptor: | Beta-arrestin 1 and single-chain fragment variable 30 (scFv30), HA signal peptide,HPC4 purification tag,Glucagon receptor,C-terminal tail of Vasopressin V2 receptor, Nanobody 32, ... | | Authors: | Chen, K, Zhang, C, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tail engagement of arrestin at the glucagon receptor.
Nature, 620, 2023
|
|
8JRV
 
 | | Cryo-EM structure of the glucagon receptor bound to glucagon and beta-arrestin 1 | | Descriptor: | Beta-arrestin 1 and single-chain fragment variable 30 (scFv30), Glucagon, HA signal peptide,HPC4 purification tag,Glucagon receptor,C-terminal tail of Vasopressin V2 receptor, ... | | Authors: | Chen, K, Zhang, C, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Tail engagement of arrestin at the glucagon receptor.
Nature, 620, 2023
|
|
7DTY
 
 | | Structural basis of ligand selectivity conferred by the human glucose-dependent insulinotropic polypeptide receptor | | Descriptor: | CHOLESTEROL, Gastric inhibitory polypeptide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, F.H, Zhang, C, Zhou, Q.T, Hang, K.N, Zou, X.Y, Chen, Y, Wu, F, Rao, Q.D, Dai, A.T, Yin, W.C, Shen, D.D, Zhang, Y, Xia, T, Stevens, R.C, Xu, H.E, Yang, D.H, Zhao, L.H, Wang, M.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-08-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into hormone recognition by the human glucose-dependent insulinotropic polypeptide receptor.
Elife, 10, 2021
|
|
8CX6
 
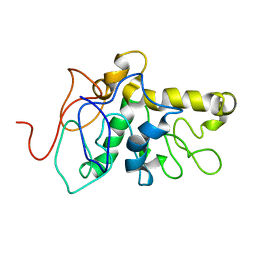 | | TPX2 Minimal Active Domain on Microtubules | | Descriptor: | Targeting protein for Xklp2-A | | Authors: | Guo, C, Alfaro-Aco, R, Russell, R, Zhang, C, Petry, S, Polenova, T. | | Deposit date: | 2022-05-19 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of protein condensation on microtubules underlying branching microtubule nucleation.
Nat Commun, 14, 2023
|
|
7Y5N
 
 | | Structure of 1:1 PAPP-A.ProMBP complex(half map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone marrow proteoglycan, ... | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-06-17 | | Release date: | 2023-01-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
7Y5Q
 
 | | Structure of 1:1 PAPP-A.STC2 complex(half map) | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-06-17 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
6IJI
 
 | | Crystal structure of PDE10 in complex with inhibitor 2b | | Descriptor: | 2-{2-[5-methyl-1-(pyridin-4-yl)-1H-benzimidazol-2-yl]ethyl}-1H-benzo[de]isoquinoline-1,3(2H)-dione, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.Y, Yu, Y.F, Zhang, C, Wu, D, Wu, Y, Luo, H.B. | | Deposit date: | 2018-10-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Validation of Phosphodiesterase-10 as a Novel Target for Pulmonary Arterial Hypertension via Highly Selective and Subnanomolar Inhibitors.
J. Med. Chem., 62, 2019
|
|
6IJH
 
 | | Crystal structure of PDE10 in complex with inhibitor AF-399/14387019 | | Descriptor: | 2-[2-(4-phenyl-5-sulfanylidene-4,5-dihydro-1H-1,2,4-triazol-3-yl)ethyl]-1H-benzo[de]isoquinoline-1,3(2H)-dione, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.Y, Yu, Y.F, Zhang, C, Wu, D, Wu, Y, Luo, H.B. | | Deposit date: | 2018-10-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Validation of Phosphodiesterase-10 as a Novel Target for Pulmonary Arterial Hypertension via Highly Selective and Subnanomolar Inhibitors.
J. Med. Chem., 62, 2019
|
|
8XTP
 
 | | Comamonas testosteroni KF-1 circularly permuted group II intron Post-2S state | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (133-MER), ... | | Authors: | Wang, L, Xie, J.H, Zhang, C, Zou, J, Huang, Z.R, Shang, S.T, Chen, X.Y, Yang, Y, Liu, J, Dong, H.H, Huang, D.M, Su, Z.M. | | Deposit date: | 2024-01-11 | | Release date: | 2025-02-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of circularly permuted group II intron self-splicing.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8XTQ
 
 | | Comamonas testosteroni KF-1 circularly permuted group II intron 1S state | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (808-MER) | | Authors: | Wang, L, Xie, J.H, Zhang, C, Zou, J, Huang, Z.R, Shang, S.T, Chen, X.Y, Yang, Y, Liu, J, Dong, H.H, Huang, D.M, Su, Z.M. | | Deposit date: | 2024-01-11 | | Release date: | 2025-02-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis of circularly permuted group II intron self-splicing.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8XTR
 
 | | Comamonas testosteroni KF-1 circularly permuted group II intron 2S state | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (219-MER), ... | | Authors: | Wang, L, Xie, J.H, Zhang, C, Zou, J, Huang, Z, Shang, S, Chen, X, Yang, Y, Liu, J, Dong, H, Huang, D, Su, Z. | | Deposit date: | 2024-01-11 | | Release date: | 2025-02-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural basis of circularly permuted group II intron self-splicing.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8XTS
 
 | | Comamonas testosteroni KF-1 circularly permuted group II intron Pre-1S state | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (814-MER) | | Authors: | Wang, L, Xie, J.H, Zhang, C, Zou, J, Huang, Z.R, Shang, S.T, Chen, X.Y, Yang, Y, Liu, J, Dong, H.H, Huang, D.M, Su, Z.M. | | Deposit date: | 2024-01-11 | | Release date: | 2025-02-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural basis of circularly permuted group II intron self-splicing.
Nat.Struct.Mol.Biol., 32, 2025
|
|
7XD7
 
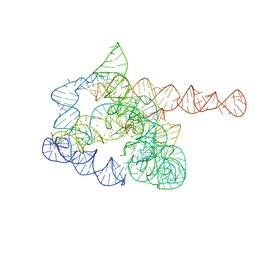 | | The pre-Tet-C state of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The pre-Tet-C state molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD3
 
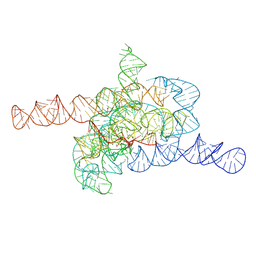 | | The relaxed pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, The relaxed pre-Tet-S1 state molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 6nt 3'/5'-exon | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD4
 
 | | The intermediate pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon | | Descriptor: | Co-transcriptional folded wild-type Tetrahymena group I intron with 6nt 3'/5'-exon, MAGNESIUM ION | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD5
 
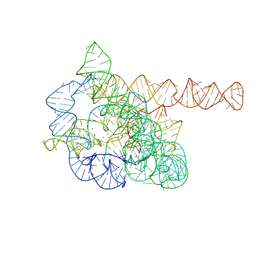 | | The Tet-S2 state of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The Tet-S2 state molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon (5'-exon), ... | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-04-05 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD6
 
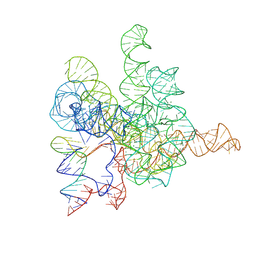 | | The Tet-S2 state with a pseudoknotted 4-way junction of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The Tet-S2 state with a pseudoknotted 4-way junction molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon (5'-exon), ... | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-04-05 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7BW1
 
 | | Crystal structure of Steroid 5-alpha-reductase 2 in complex with Finasteride | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-oxo-5-alpha-steroid 4-dehydrogenase 2, SULFATE ION, ... | | Authors: | Xiao, Q, Zhang, C, Wei, Z. | | Deposit date: | 2020-04-13 | | Release date: | 2020-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human steroid 5 alpha-reductase 2 with anti-androgen drug finasteride.
Res Sq, 2020
|
|
