9GCX
 
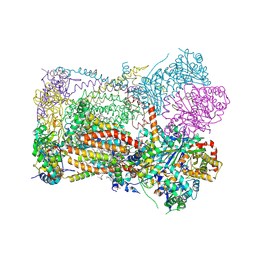 | | Crystal structure of bovine Cytochrome bc1 in complex with inhibitor F8 | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, 2-methyl-3-(4-(4-(trifluoromethoxy)phenoxy)phenyl)-1,5,7,8-tetrahydro-4H-pyrano[4,3-b]pyridin-4-one, ... | | Authors: | Pinthong, N, Hong, W, ONeill, P.W, Antonyuk, S.V. | | Deposit date: | 2024-08-03 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (3.522 Å) | | Cite: | Novel antimalarial 3-substituted quinolones isosteres with improved pharmacokinetic properties.
Eur.J.Med.Chem., 284, 2024
|
|
9J0H
 
 | |
2HFW
 
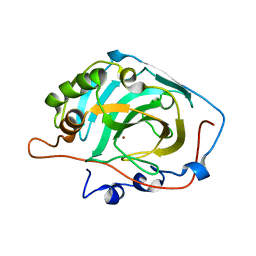 | | Structural and kinetic analysis of proton shuttle residues in the active site of human carbonic anhydrase III | | Descriptor: | Carbonic anhydrase 3, ZINC ION | | Authors: | Elder, I, Fisher, S.Z, Laipis, P.J, Tu, C.K, McKenna, R, Silverman, D.N. | | Deposit date: | 2006-06-26 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and kinetic analysis of proton shuttle residues in the active site of human carbonic anhydrase III.
Proteins, 68, 2007
|
|
7LQV
 
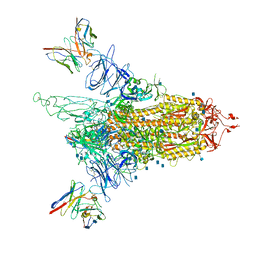 | | Cryo-EM structure of NTD-directed neutralizing antibody 4-8 Fab in complex with SARS-CoV-2 S2P spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-8 Heavy Chain, 4-8 Light chain, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Potent SARS-CoV-2 Neutralizing Antibodies Directed Against Spike N-Terminal Domain Target a Single Supersite
Cell Host Microbe, 2021
|
|
7LQW
 
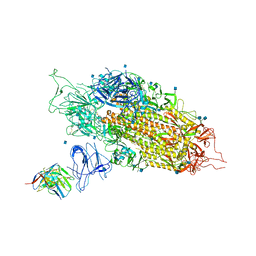 | | Cryo-EM structure of NTD-directed neutralizing antibody 2-17 Fab in complex with SARS-CoV-2 S2P spike | | Descriptor: | 2-17 Heavy Chain, 2-17 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-24 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite.
Cell Host Microbe, 29, 2021
|
|
7L2E
 
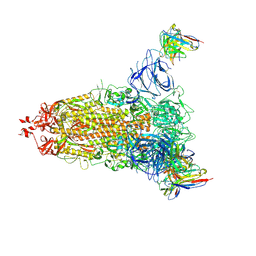 | |
7L2F
 
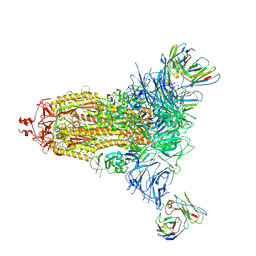 | |
7L2D
 
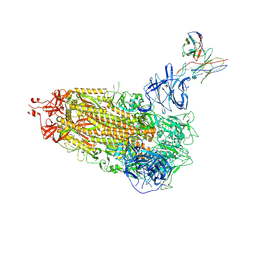 | |
7L57
 
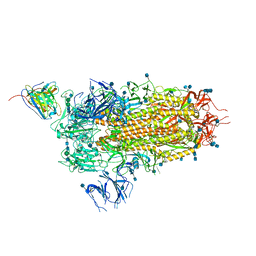 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rapp, M, Shapiro, L. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.87 Å) | | Cite: | Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class.
Cell Rep, 35, 2021
|
|
7L58
 
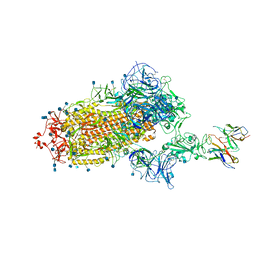 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab H4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab H4 variable domain heavy chain, ... | | Authors: | Rapp, M, Shapiro, L. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.07 Å) | | Cite: | Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class.
Cell Rep, 35, 2021
|
|
7L56
 
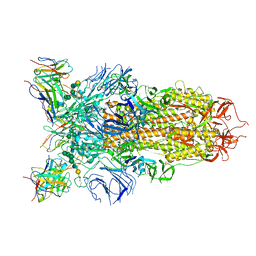 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2-43 variable domain heavy chain, ... | | Authors: | Rapp, M, Shapiro, L. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class.
Cell Rep, 35, 2021
|
|
5CFE
 
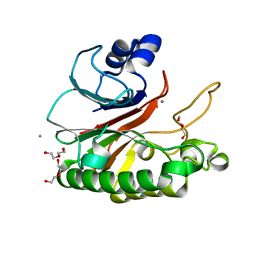 | | Bacillus subtilis AP endonuclease ExoA | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Exodeoxyribonuclease | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2015-07-08 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural comparison of AP endonucleases from the exonuclease III family reveals new amino acid residues in human AP endonuclease 1 that are involved in incision of damaged DNA.
Biochimie, 128-129, 2016
|
|
7L2C
 
 | | Crystallographic structure of neutralizing antibody 2-51 in complex with SARS-CoV-2 spike N-terminal domain (NTD) | | Descriptor: | 2-51 heavy chain, 2-51 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cerutti, G, Reddem, E.R, Shapiro, L. | | Deposit date: | 2020-12-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite.
Cell Host Microbe, 29, 2021
|
|
5CFG
 
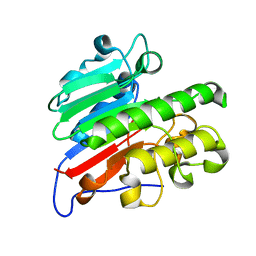 | | C2 crystal form of APE1 with Mg2+ | | Descriptor: | DNA-(apurinic or apyrimidinic site) lyase, MAGNESIUM ION | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2015-07-08 | | Release date: | 2016-07-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural comparison of AP endonucleases from the exonuclease III family reveals new amino acid residues in human AP endonuclease 1 that are involved in incision of damaged DNA.
Biochimie, 128-129, 2016
|
|
8TWX
 
 | | Synthesis and Evaluation of Diaryl Ether Modulators of the Leukotriene A4 Hydrolase Aminopeptidase Activity | | Descriptor: | 5-[4-(4-chlorophenoxy)phenyl]-1H-pyrazol-3-amine, Leukotriene A-4 hydrolase, ZINC ION | | Authors: | Lee, K.H, Lee, S.H, Paige, M, Noble, S.M. | | Deposit date: | 2023-08-21 | | Release date: | 2024-08-28 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthesis and Evaluation of diaryl ether modulators of the leukotriene A 4 hydrolase aminopeptidase activity.
Eur.J.Med.Chem., 272, 2024
|
|
4S1R
 
 | | Crystal structure of a VRC01-lineage antibody, 45-VRC01.H08.F-117225, in complex with clade A/E HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab of VRC01 light chain, Fab of VRC01-lineage antibody,45-VRC01.H08.F-117225 heavy chain, ... | | Authors: | Kwon, Y.D, Yang, Y, Zhang, B, Kwong, P.D. | | Deposit date: | 2015-01-14 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell(Cambridge,Mass.), 161, 2015
|
|
4S1Q
 
 | | Crystal structure of a VRC01-lineage antibody, 45-VRC01.H03+06.D-001739, in complex with clade A/E HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab of VRC01 light chain, Fab of VRC01-lineage antibody,45-VRC01.H03+06.D-001739 heavy chain, ... | | Authors: | Kwon, Y.D, Yang, Y, Zhang, B, Kwong, P.D. | | Deposit date: | 2015-01-14 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell(Cambridge,Mass.), 161, 2015
|
|
4S1S
 
 | | Crystal structure of a VRC01-lineage antibody, 45-VRC01.H5.F-185917, in complex with clade A/E HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fab of VRC01 light chain, ... | | Authors: | Kwon, Y.D, Yang, Y, Zhang, B, Kwong, P.D. | | Deposit date: | 2015-01-14 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell(Cambridge,Mass.), 161, 2015
|
|
7CVN
 
 | | The N-arylsulfonyl-indole-2-carboxamide-based inhibitors against fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4-(3-acetamidophenyl)-N-(4-methoxyphenyl)sulfonyl-7-nitro-1H-indole-2-carboxamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X, Zhou, J, Xu, B. | | Deposit date: | 2020-08-26 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Design,synthesis,biological evaluation and binding mode analysis of 7-nitro-indole-N-acylarylsulfonamide-based fructose-1,6-bisphosphatase inhibitors
Chinese journal of medicinal chemistry, 30, 2020
|
|
6NZ7
 
 | |
4PMH
 
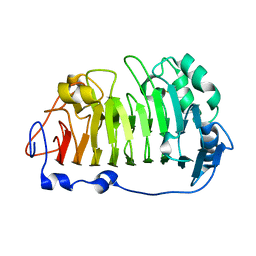 | | The structure of rice weevil pectin methyl esterase | | Descriptor: | Pectinesterase | | Authors: | Stenkamp, R.E, Teller, D.C, Behnke, C.A, Reeck, G.R. | | Deposit date: | 2014-05-21 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The structure of rice weevil pectin methylesterase.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4QE5
 
 | |
4QE4
 
 | |
4QEH
 
 | |
4DVO
 
 | | Room-temperature joint X-ray/neutron structure of D-xylose isomerase in complex with 2Ni2+ and per-deuterated D-sorbitol at pH 5.9 | | Descriptor: | NICKEL (II) ION, Xylose isomerase, sorbitol | | Authors: | Kovalevsky, A.Y, Hanson, L, Langan, P. | | Deposit date: | 2012-02-23 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Inhibition of D-xylose isomerase by polyols: atomic details by joint X-ray/neutron crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
