6H6G
 
 | | Crystal Structure of TcdB2-TccC3 without hypervariable C-terminal region | | Descriptor: | TcdB2,TccC3 | | Authors: | Gatsogiannis, C, Merino, F, Roderer, D, Balchin, D, Schubert, E, Kuhlee, A, Hayer-Hartl, M, Raunser, S. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Tc toxin activation requires unfolding and refolding of a beta-propeller.
Nature, 563, 2018
|
|
5YYC
 
 | | Crystal structure of alanine racemase from Bacillus pseudofirmus (OF4) | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dong, H, Hu, T.T, He, G.Z, Lu, D.R, Qi, J.X, Dou, Y.S, Long, W, He, X, Su, D, Ju, J.S. | | Deposit date: | 2017-12-08 | | Release date: | 2019-01-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural features and kinetic characterization of alanine racemase from Bacillus pseudofirmus OF4.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
5FBN
 
 | | BTK kinase domain with inhibitor 1 | | Descriptor: | 1,2-ETHANEDIOL, 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-[8-azanyl-3-[(3~{R})-1-(3-methyloxetan-3-yl)carbonylpiperidin-3-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-[4-(trifluoromethyl)pyridin-2-yl]benzamide, ... | | Authors: | Raaijmakers, H.C.A, Vu-Pham, D. | | Deposit date: | 2015-12-14 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of 8-Amino-imidazo[1,5-a]pyrazines as Reversible BTK Inhibitors for the Treatment of Rheumatoid Arthritis.
Acs Med.Chem.Lett., 7, 2016
|
|
6H67
 
 | | Yeast RNA polymerase I elongation complex stalled by cyclobutane pyrimidine dimer (CPD) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Sanz-Murillo, M, Xu, J, Gil-Carton, D, Wang, D, Fernandez-Tornero, C. | | Deposit date: | 2018-07-26 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of RNA polymerase I stalling at UV light-induced DNA damage.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2AK5
 
 | | beta PIX-SH3 complexed with a Cbl-b peptide | | Descriptor: | 8-residue peptide from a signal transduction protein CBL-B, Rho guanine nucleotide exchange factor 7 | | Authors: | Jozic, D, Cardenes, N, Deribe, Y.L, Moncalian, G, Hoeller, D, Groemping, Y, Dikic, I, Rittinger, K, Bravo, J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cbl promotes clustering of endocytic adaptor proteins.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2JPU
 
 | | solution structure of NESG target SsR10, Orf c02003 protein | | Descriptor: | Orf c02003 protein | | Authors: | Wu, Y, Singarapu, K.K, Zhang, Q, Eletski, A, Xu, D, Sukumaran, D, Parish, D, Wang, D, Jiang, M, Cunningham, K, Maglaqui, M, Owens, L, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T, Rost, B, Montelione, G, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-05-23 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of NESG target SsR10, Orf c02003 protein
To be Published
|
|
6H68
 
 | | Yeast RNA polymerase I elongation complex stalled by cyclobutane pyrimidine dimer (CPD) with fully-ordered A49 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Sanz-Murillo, M, Xu, J, Gil-Carton, D, Wang, D, Fernandez-Tornero, C. | | Deposit date: | 2018-07-26 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of RNA polymerase I stalling at UV light-induced DNA damage.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8E7O
 
 | | CRYSTAL STRUCTURE OF LYS48-LINKED TETRAUBIQUITIN | | Descriptor: | SULFATE ION, Ubiquitin | | Authors: | Lemma, B.E, Fushman, D. | | Deposit date: | 2022-08-24 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of selective recognition of Lys48-linked polyubiquitin by macrocyclic peptide inhibitors of proteasomal degradation.
Nat Commun, 14, 2023
|
|
1UPU
 
 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE, MUTANT C128V, BOUND TO PRODUCT URIDINE-1-MONOPHOSPHATE (UMP) | | Descriptor: | PHOSPHATE ION, URACIL PHOSPHORIBOSYLTRANSFERASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-04-16 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
6H6E
 
 | | PTC3 holotoxin complex from Photorhabdus luminecens in prepore state (TcdA1, TcdB2, TccC3) | | Descriptor: | TcdA1, TcdB2,TccC3 | | Authors: | Gatsogiannis, C, Merino, F, Roderer, D, Balchin, D, Schubert, E, Kuhlee, A, Hayer-Hartl, M, Raunser, S. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Tc toxin activation requires unfolding and refolding of a beta-propeller.
Nature, 563, 2018
|
|
5D3S
 
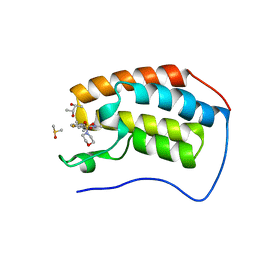 | | First bromodomain of BRD4 bound to inhibitor XD44 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-acetyl-3-ethyl-N-[4-fluoro-3-(morpholin-4-ylsulfonyl)phenyl]-5-methyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4, ... | | Authors: | Wohlwend, D, Huegle, M. | | Deposit date: | 2015-08-06 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
5D24
 
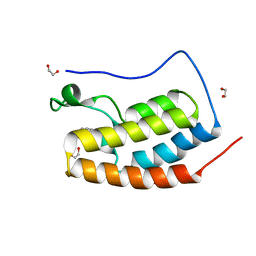 | | First bromodomain of BRD4 bound to inhibitor XD26 | | Descriptor: | 1,2-ETHANEDIOL, 4-acetyl-N-[3-(2-amino-2-oxoethoxy)phenyl]-3-ethyl-5-methyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Huegle, M, Gerhardt, S. | | Deposit date: | 2015-08-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
5D3H
 
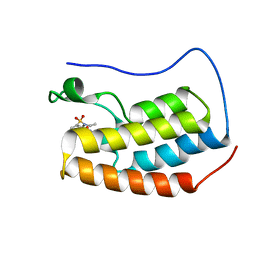 | | First bromodomain of BRD4 bound to inhibitor XD29 | | Descriptor: | Bromodomain-containing protein 4, N-[5-(diethylsulfamoyl)-2-hydroxyphenyl]-3-ethyl-4-(hydroxyacetyl)-5-methyl-1H-pyrrole-2-carboxamide | | Authors: | Wohlwend, D, Huegle, M, Weitzel, G. | | Deposit date: | 2015-08-06 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
2XGL
 
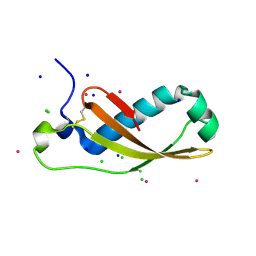 | | The X-ray structure of the Escherichia coli colicin M immunity protein demonstrates the presence of a disulphide bridge, which is functionally essential | | Descriptor: | CADMIUM ION, CHLORIDE ION, COLICIN-M IMMUNITY PROTEIN, ... | | Authors: | Gerard, F, Brooks, M.A, Barreteau, H, Touze, T, Graille, M, Bouhss, A, Blanot, D, Tilbeurgh, H.v, Mengin-Lecreulx, D. | | Deposit date: | 2010-06-07 | | Release date: | 2010-11-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-Ray Structure and Site-Directed Mutagenesis Analysis of the Escherichia Coli Colicin M Immunity Protein.
J.Bacteriol., 193, 2011
|
|
5ARF
 
 | | SMYD2 in complex with small molecule inhibitor compound-2 | | Descriptor: | N-LYSINE METHYLTRANSFERASE SMYD2, N-[3-(4-CHLOROPHENYL)-1-{N'-CYANO-N-[3-(DIFLUOROMETHOXY)PHENYL]CARBAMIMIDOYL}-4,5-DIHYDRO-1H- PYRAZOL-4-YL]-N-ETHYL-2-HYDROXYACETAMIDE, S-ADENOSYLMETHIONINE, ... | | Authors: | Hillig, R.C, Badock, V, Barak, N, Stellfeld, T, Eggert, E, ter Laak, A, Weiske, J, Christ, C.D, Koehr, S, Stoeckigt, D, Mowat, J, Mueller, T, Fernandez-Montalvan, A.E, Hartung, I.V, Stresemann, C, Brumby, T, Weinmann, H. | | Deposit date: | 2015-09-24 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery and Characterization of a Highly Potent and Selective Aminopyrazoline-Based in Vivo Probe (Bay-598) for the Protein Lysine Methyltransferase Smyd2.
J.Med.Chem., 59, 2016
|
|
4ABI
 
 | | Co-complex structure of bovine trypsin with a modified Bowman-Birk inhibitor (PtA)SFTI-1(1,14), that was 1,4-disubstituted with a 1,2,3- trizol to mimic a trans amide bond | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, DIMETHYLFORMAMIDE, ... | | Authors: | Schmelz, S, Empting, M, Tischler, M, Nasu, D, Heinz, D, Kolmar, H. | | Deposit date: | 2011-12-08 | | Release date: | 2012-03-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Braces for the Peptide Backbone: Insights Into Structure-Activity Relation-Ships of Protease Inhibitor Mimics with Locked Amide Conformations
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
2FOE
 
 | | Structure of porcine pancreatic elastase in 80% hexane | | Descriptor: | (2S)-HEX-5-ENE-1,2-DIOL, CALCIUM ION, SULFATE ION, ... | | Authors: | Mattos, C, Bellamacina, C.R, Peisach, E, Pereira, A, Vitkup, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple solvent crystal structures: Probing binding sites, plasticity and hydration
J.Mol.Biol., 357, 2006
|
|
8WIU
 
 | | Bromodomain and Extra-terminal Domain (BET) BRD4 | | Descriptor: | 7-[5-[1-(cyclopropylmethyl)-3,5-dimethyl-pyrazol-4-yl]pyridin-3-yl]-1~{H}-imidazo[4,5-b]pyridine, Isoform C of Bromodomain-containing protein 4 | | Authors: | Cao, D, Zhiyan, D, Xiong, B. | | Deposit date: | 2023-09-25 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of a brain-permeable bromodomain and extra terminal domain (BET) inhibitor with selectivity for BD1 for the treatment of multiple sclerosis.
Eur.J.Med.Chem., 265, 2023
|
|
2ZBK
 
 | | Crystal structure of an intact type II DNA topoisomerase: insights into DNA transfer mechanisms | | Descriptor: | RADICICOL, Type 2 DNA topoisomerase 6 subunit B, Type II DNA topoisomerase VI subunit A | | Authors: | Graille, M, Cladiere, L, Durand, D, Lecointe, F, Forterre, P, van Tilbeurgh, H, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2007-10-22 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Crystal Structure of an Intact Type II DNA Topoisomerase: Insights into DNA Transfer Mechanisms
Structure, 16, 2008
|
|
6HPV
 
 | | Crystal structure of mouse fetuin-B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Fetuin-B | | Authors: | Fahrenkamp, D, Dietzel, E, de Sanctis, D, Jovine, L. | | Deposit date: | 2018-09-22 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of mammalian plasma fetuin-B and its mechanism of selective metallopeptidase inhibition.
Iucrj, 6, 2019
|
|
2FOC
 
 | | Structure of porcine pancreatic elastase in 55% dimethylformamide | | Descriptor: | CALCIUM ION, DIMETHYLFORMAMIDE, SULFATE ION, ... | | Authors: | Mattos, C, Bellamacina, C.R, Peisach, E, Pereira, A, Vitkup, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiple solvent crystal structures: Probing binding sites, plasticity and hydration
J.Mol.Biol., 357, 2006
|
|
3GLM
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with crotonyl-coA | | Descriptor: | CHLORIDE ION, CROTONYL COENZYME A, Glutaconyl-CoA decarboxylase subunit A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-03-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
8TXO
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound to the unnatural dZ-PTP base pair in the active site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Aiyer, S, Shan, Z, Oh, J, Wang, D, Tan, Y.Z, Lyumkis, D. | | Deposit date: | 2023-08-23 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Overcoming resolution attenuation during tilted cryo-EM data collection.
Nat Commun, 15, 2024
|
|
6AXL
 
 | | Crystal structure of Fab317 complex | | Descriptor: | Fab317 heavy chain, Fab317 light chains, Peptide ACE-ASN-PRO-ASN-ALA-ASN-PRO-ASN-ALA-ASN-PRO-ASN-ALA-NH2 | | Authors: | Oyen, D, Wilson, I.A. | | Deposit date: | 2017-09-07 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for antibody recognition of the NANP repeats in Plasmodium falciparum circumsporozoite protein.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1XLY
 
 | | X-RAY STRUCTURE OF THE RNA-BINDING PROTEIN SHE2p | | Descriptor: | SHE2p | | Authors: | Niessing, D, Huettelmaier, S, Zenklusen, D, Singer, R.H, Burley, S.K. | | Deposit date: | 2004-09-30 | | Release date: | 2004-11-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | She2p is a novel RNA binding protein with a basic helical hairpin motif
Cell(Cambridge,Mass.), 119, 2004
|
|
