3OHS
 
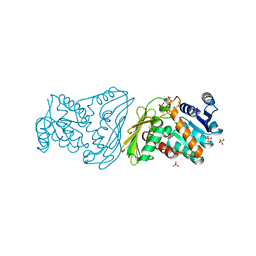 | |
2O4U
 
 | | Crystal structure of Mammalian Dimeric Dihydrodiol Dehydrogenase | | Descriptor: | BETA-MERCAPTOETHANOL, Dimeric dihydrodiol dehydrogenase, GLYCEROL, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2006-12-04 | | Release date: | 2007-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of dimeric dihydrodiol dehydrogenase apoenzyme and inhibitor complex: probing the subunit interface with site-directed mutagenesis.
Proteins, 70, 2008
|
|
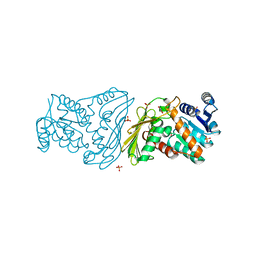
3C3U
 
 | | Crystal structure of AKR1C1 in complex with NADP and 3,5-dichlorosalicylic acid | | Descriptor: | 3,5-dichloro-2-hydroxybenzoic acid, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Dhagat, U, El-Kabbani, O. | | Deposit date: | 2008-01-28 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selectivity determinants of inhibitor binding to human 20alpha-hydroxysteroid dehydrogenase: crystal structure of the enzyme in ternary complex with coenzyme and the potent inhibitor 3,5-dichlorosalicylic acid
J.Med.Chem., 51, 2008
|
|
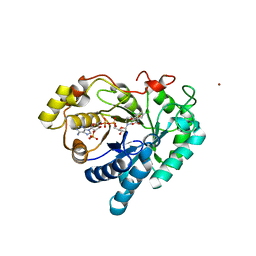
3CV6
 
 | | The crystal structure of mouse 17-alpha hydroxysteroid dehydrogenase GG225.226PP mutant in complex with inhibitor and cofactor NADP+. | | Descriptor: | 4-[(1R,2S)-1-ethyl-2-(4-hydroxyphenyl)butyl]phenol, Aldo-keto reductase family 1 member C21, BETA-MERCAPTOETHANOL, ... | | Authors: | Dhagat, U, El-Kabbani, O. | | Deposit date: | 2008-04-17 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the G225P/G226P mutant of mouse 3(17)alpha-hydroxysteroid dehydrogenase (AKR1C21) ternary complex: implications for the binding of inhibitor and substrate.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
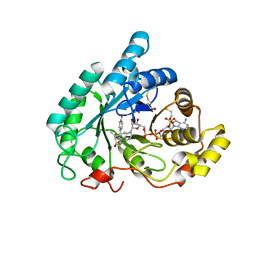
3D3W
 
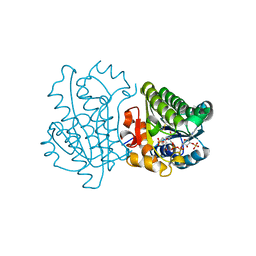 | |
3CV7
 
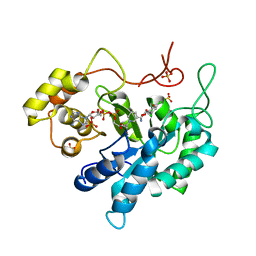 | | Crystal structure of porcine aldehyde reductase ternary complex | | Descriptor: | 3,5-dichloro-2-hydroxybenzoic acid, Alcohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2008-04-18 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Structure of aldehyde reductase in ternary complex with coenzyme and the potent 20alpha-hydroxysteroid dehydrogenase inhibitor 3,5-dichlorosalicylic acid: Implications for inhibitor binding and selectivity
Arch.Biochem.Biophys., 479, 2008
|
|
7NUG
 
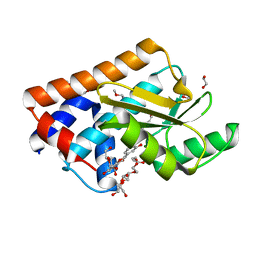 | |
7NUH
 
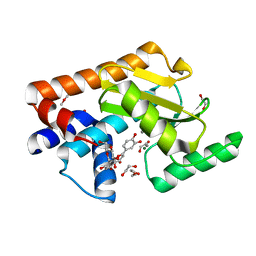 | |
2D1Y
 
 | |
3FX4
 
 | | Porcine aldehyde reductase in ternary complex with inhibitor | | Descriptor: | Alcohol dehydrogenase [NADP+], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2009-01-20 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of aldehyde reductase in ternary complex with a 5-arylidene-2,4-thiazolidinedione aldose reductase inhibitor.
Eur.J.Med.Chem., 45, 2010
|
|
3FJN
 
 | |
3GUG
 
 | |
1JQN
 
 | |
1JQO
 
 | |
2IHY
 
 | | Structure of the Staphylococcus aureus putative ATPase subunit of an ATP-binding cassette (ABC) transporter | | Descriptor: | ABC transporter, ATP-binding protein, SULFATE ION | | Authors: | McGrath, T.E, Yu, C.S, Romanov, V, Lam, R, Dharamsi, A, Virag, C, Mansoury, K, Thambipillai, D, Richards, D, Guthrie, J, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2006-09-27 | | Release date: | 2007-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Staphylococcus aureus putative ATPase subunit of an ATP-binding cassette (ABC) transporter
To be Published
|
|
4TNO
 
 | | Hypothetical protein PF1117 from Pyrococcus Furiosus: Structure solved by sulfur-SAD using Swiss Light Source Data | | Descriptor: | CHLORIDE ION, CRISPR-associated endoribonuclease Cas2 | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-06-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
5X7X
 
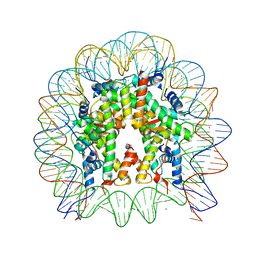 | | The crystal structure of the nucleosome containing H3.3 at 2.18 angstrom resolution | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Arimura, Y, Taguchi, H, Kurumizaka, H. | | Deposit date: | 2017-02-27 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.184 Å) | | Cite: | Crystal Structure and Characterization of Novel Human Histone H3 Variants, H3.6, H3.7, and H3.8
Biochemistry, 56, 2017
|
|
6OU3
 
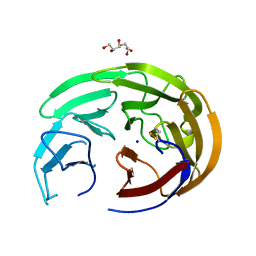 | |
6OU2
 
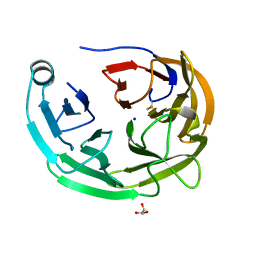 | |
6OU1
 
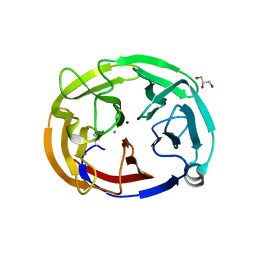 | | Crystal Structure of the Computationally-derived 21-Variant of the Myocilin Olfactomedin Domain | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Hill, S.E, Kwon, M.S, Lieberman, R.L. | | Deposit date: | 2019-05-03 | | Release date: | 2019-07-03 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Stable calcium-free myocilin olfactomedin domain variants reveal challenges in differentiating between benign and glaucoma-causing mutations.
J.Biol.Chem., 294, 2019
|
|
6OU0
 
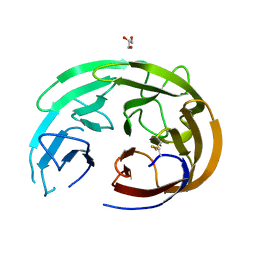 | |
7AOT
 
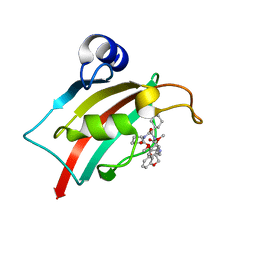 | | The Fk1 domain of FKBP51 in complex with (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0-5,10]tetracosa- 1(24),20,22-triene-4,11,14,17-tetrone | | Descriptor: | (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0-5,10]tetracosa- 1(24),20,22-triene-4,11,14,17-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, A.M, Meyners, C, Heymann, T, Merz, S, Purder, P, Bracher, A, Hausch, F. | | Deposit date: | 2020-10-15 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Macrocyclic FKBP51 Ligands Define a Transient Binding Mode with Enhanced Selectivity.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7WVL
 
 | |
4WAU
 
 | | Crystal structure of CENP-M solved by native-SAD phasing | | Descriptor: | Centromere protein M | | Authors: | Weinert, T, Basilico, F, Cecatiello, V, Pasqualato, S, Wang, M. | | Deposit date: | 2014-09-01 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4WAB
 
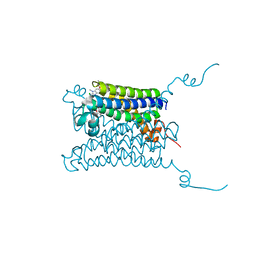 | | Crystal structure of mPGES1 solved by native-SAD phasing | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-[(2,2-dimethylpropanoylamino)methyl]phenyl]amino]-1-methyl-6-(2-methyl-2-oxidanyl-propoxy)-N-[2,2,2-tris(fluoranyl)ethyl]benzimidazole-5-carboxamide, GLUTATHIONE, Prostaglandin E synthase,Leukotriene C4 synthase | | Authors: | Weinert, T, Li, D, Howe, N, Caffrey, M, Wang, M. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
