5NN9
 
 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | Deposit date: | 1991-03-28 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
6P2B
 
 | | Tethered PXR-LBD/SRC-1p bound to Garcinoic Acid | | Descriptor: | (2Z,6E,10E)-13-[(2R)-6-hydroxy-2,8-dimethyl-3,4-dihydro-2H-1-benzopyran-2-yl]-2,6,10-trimethyltrideca-2,6,10-trienoic acid, DIMETHYL SULFOXIDE, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2019-05-21 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Garcinoic Acid Is a Natural and Selective Agonist of Pregnane X Receptor.
J.Med.Chem., 63, 2020
|
|
3K7N
 
 | | Structures of two elapid snake venom metalloproteases with distinct activities highlight the disulfide patterns in the D domain of ADAMalysin family proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Guan, H.H, Huang, Y.W, Wu, W.G, Chen, C.J. | | Deposit date: | 2009-10-13 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of two elapid snake venom metalloproteases with distinct activities highlight the disulfide patterns in the D domain of ADAMalysin family proteins
J.Struct.Biol., 169, 2010
|
|
6NN9
 
 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | Deposit date: | 1991-03-28 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
1HC1
 
 | |
5TIM
 
 | | REFINED 1.83 ANGSTROMS STRUCTURE OF TRYPANOSOMAL TRIOSEPHOSPHATE ISOMERASE, CRYSTALLIZED IN THE PRESENCE OF 2.4 M-AMMONIUM SULPHATE. A COMPARISON WITH THE STRUCTURE OF THE TRYPANOSOMAL TRIOSEPHOSPHATE ISOMERASE-GLYCEROL-3-PHOSPHATE COMPLEX | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Wierenga, R.K, Hol, W.G.J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Refined 1.83 A structure of trypanosomal triosephosphate isomerase crystallized in the presence of 2.4 M-ammonium sulphate. A comparison with the structure of the trypanosomal triosephosphate isomerase-glycerol-3-phosphate complex.
J.Mol.Biol., 220, 1991
|
|
1QS0
 
 | | Crystal Structure of Pseudomonas Putida 2-oxoisovalerate Dehydrogenase (Branched-Chain Alpha-Keto Acid Dehydrogenase, E1B) | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, 2-OXOISOVALERATE DEHYDROGENASE ALPHA-SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA-SUBUNIT, ... | | Authors: | Aevarsson, A, Seger, K, Turley, S, Sokatch, J.R, Hol, W.G.J. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-18 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of 2-oxoisovalerate and dehydrogenase and the architecture of 2-oxo acid dehydrogenase multienzyme complexes.
Nat.Struct.Biol., 6, 1999
|
|
7AF2
 
 | | Salmonella typhimurium neuraminidase mutant (D62G) | | Descriptor: | GLYCEROL, PHOSPHATE ION, Sialidase | | Authors: | Salinger, M.T, Kuhn, P, Laver, W.G, Pape, T, Schneider, T.R, Sheldrick, G.M, Vimr, E.R, Garman, E.F. | | Deposit date: | 2020-09-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.792 Å) | | Cite: | Salmonella typhimurium neuraminidase mutant (D62G)
To Be Published
|
|
7AEY
 
 | | Salmonella typhimurium neuraminidase in complex with isocarba-DANA. | | Descriptor: | (3~{S},4~{S},5~{R})-4-acetamido-3-oxidanyl-5-[(1~{S},2~{R})-1,2,3-tris(oxidanyl)propyl]cyclohexane-1-carboxylic acid, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Salinger, M.T, Kuhn, P, Laver, W.G, Pape, T, Schneider, T.R, Sheldrick, G.M, Vasella, A.T, Vimr, E.R, Vorwerk, S, Garman, E.F. | | Deposit date: | 2020-09-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.919 Å) | | Cite: | Salmonella typhimurium neuraminidase in complex with isocarba-DANA.
To Be Published
|
|
3NN9
 
 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | Deposit date: | 1991-03-28 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
6QB8
 
 | | Human CCT:mLST8 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Cuellar, J, Santiago, C, Ludlam, W.G, Bueno-Carrasco, M.T, Valpuesta, J.M, Willardson, B.M. | | Deposit date: | 2018-12-20 | | Release date: | 2019-07-03 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural and functional analysis of the role of the chaperonin CCT in mTOR complex assembly.
Nat Commun, 10, 2019
|
|
5J58
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor (Chem 1856) | | Descriptor: | (3S)-1-(5-chloro-1H-imidazo[4,5-b]pyridin-2-yl)-N-[(3,5-dichlorophenyl)methyl]piperidin-3-amine, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Barros-Alvarez, X, Hol, W.G.J. | | Deposit date: | 2016-04-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-guided design of novel Trypanosoma brucei Methionyl-tRNA synthetase inhibitors.
Eur J Med Chem, 124, 2016
|
|
5J59
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor (Chem 1893) | | Descriptor: | 5-chloro-N-(2-{[(3,5-dichlorophenyl)methyl]amino}ethyl)-N-methyl-3H-imidazo[4,5-b]pyridin-2-amine, GLYCEROL, METHIONINE, ... | | Authors: | Barros-Alvarez, X, Hol, W.G.J. | | Deposit date: | 2016-04-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-guided design of novel Trypanosoma brucei Methionyl-tRNA synthetase inhibitors.
Eur J Med Chem, 124, 2016
|
|
359D
 
 | |
4CSB
 
 | | Structure of the Virulence-Associated Protein VapD from the intracellular pathogen Rhodococcus equi. | | Descriptor: | VIRULENCE ASSOCIATED PROTEIN VAPD, octyl beta-D-glucopyranoside | | Authors: | Whittingham, J.L, Blagova, E.V, Finn, C.E, Luo, H, Miranda-CasoLuengo, R, Turkenburg, J.P, Leech, A.P, Walton, P.H, Meijers, W.G, Wilkinson, A.J. | | Deposit date: | 2014-03-06 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Virulence-Associated Protein Vapd from the Intracellular Pathogen Rhodococcus Equi.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3BSQ
 
 | | Crystal structure of human kallikrein 7 produced as a secretion protein in E.coli | | Descriptor: | Kallikrein-7, SULFATE ION | | Authors: | Fernandez, I.S, Standker, L, Magert, H.J, Forssmann, W.G, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2007-12-26 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human epidermal kallikrein 7 (hK7) synthesized directly in its native state in E. coli: insights into the atomic basis of its inhibition by LEKTI domain 6 (LD6)
J.Mol.Biol., 377, 2008
|
|
7RZI
 
 | | Insulin Degrading Enzyme pC/pC | | Descriptor: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZE
 
 | | Insulin Degrading Enzyme pO/pC | | Descriptor: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZH
 
 | | Insulin Degrading Enzyme O/O | | Descriptor: | Cysteine-free Insulin-degrading enzyme | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZF
 
 | | Insulin Degrading Enzyme O/pC | | Descriptor: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZG
 
 | | Insulin Degrading Enzyme O/pO | | Descriptor: | Cysteine-free Insulin-degrading enzyme | | Authors: | Mancl, J.M, Liang, W.G, Tang, W.J. | | Deposit date: | 2021-08-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
4MW2
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-(3-{[5-chloro-2-hydroxy-3-(prop-2-en-1-yl)benzyl]amino}propyl)-3-thiophen-3-ylurea (Chem 1472) | | Descriptor: | 1-(3-{[5-chloro-2-hydroxy-3-(prop-2-en-1-yl)benzyl]amino}propyl)-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4MW7
 
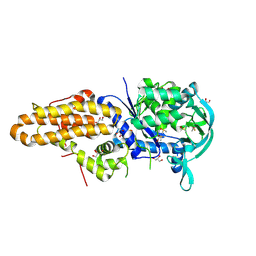 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(5-chloro-2-ethoxy-3-iodobenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1469) | | Descriptor: | 1-{3-[(5-chloro-2-ethoxy-3-iodobenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
3CHB
 
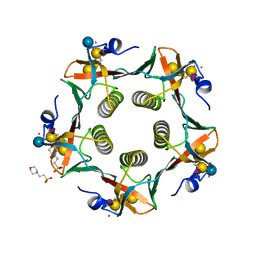 | | CHOLERA TOXIN B-PENTAMER COMPLEXED WITH GM1 PENTASACCHARIDE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHOLERA TOXIN, UNKNOWN ATOM OR ION, ... | | Authors: | Merritt, E.A, Hol, W.G.J. | | Deposit date: | 1998-03-24 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The 1.25 A resolution refinement of the cholera toxin B-pentamer: evidence of peptide backbone strain at the receptor-binding site.
J.Mol.Biol., 282, 1998
|
|
5COR
 
 | |
